Proximal glycans outside of the epitopes regulate the presentation of HIV-1 envelope gp120 helper epitopes
- PMID: 19414790
- PMCID: PMC2808118
- DOI: 10.4049/jimmunol.0804287
Proximal glycans outside of the epitopes regulate the presentation of HIV-1 envelope gp120 helper epitopes
Abstract
Glycosylation of HIV-1 envelope gp120 determines not only the proper structure, but also the immune responses against this Ag. Although glycans may be part of specific epitopes or shield other epitopes from T cells and Abs, this study provides evidence for a different immunomodulatory function of glycans associated with gp120 residues N230 and N448. These glycans are required for efficient MHC class II-restricted presentation of nearby CD4 T cell epitopes, even though they are not part of the epitopes. The glycans do not affect CD4 T cell recognition of more distant epitopes and are not essential for the proper folding and function of gp120. Data on CD4 T cell recognition of N448 mutants combined with proteolysis analyses and surface electrostatic potential calculation around residue N448 support the notion that N448 glycan near the epitope's C terminus renders the site to be surface accessible and allows its efficient processing. In contrast, the N230 glycan contributes to the nearby epitope presentation at a step other than the proteolytic processing of the epitope. Hence, N-glycans can determine CD4 T cell recognition of nearby gp120 epitopes by regulating the different steps in the MHC class II processing and presentation pathway after APCs acquire the intact gp120 Ag exogenously. Modifications of amino acids bearing glycans at the C termini of gp120 helper epitopes may prove to be a useful strategy for enhancing the immunogenicity of HIV-1 envelope gp120.
Figures

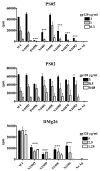

References
-
- Zhang M, Gaschen B, Blay W, Foley B, Haigwood N, Kuiken C, Korber B. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology. 2004;14:1229–1246. - PubMed
-
- Morikawa Y, Moore JP, Jones IM. HIV-1 envelope protein gp120 expression by secretion in E. coli: assessment of CD4 binding and use in epitope mapping. J Virol Methods. 1990;29:105–113. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

