How large is the metabolome? A critical analysis of data exchange practices in chemistry
- PMID: 19415114
- PMCID: PMC2673031
- DOI: 10.1371/journal.pone.0005440
How large is the metabolome? A critical analysis of data exchange practices in chemistry
Abstract
Background: Calculating the metabolome size of species by genome-guided reconstruction of metabolic pathways misses all products from orphan genes and from enzymes lacking annotated genes. Hence, metabolomes need to be determined experimentally. Annotations by mass spectrometry would greatly benefit if peer-reviewed public databases could be queried to compile target lists of structures that already have been reported for a given species. We detail current obstacles to compile such a knowledge base of metabolites.
Results: As an example, results are presented for rice. Two rice (oryza sativa) subspecies have been fully sequenced, oryza japonica and oryza indica. Several major small molecule databases were compared for listing known rice metabolites comprising PubChem, Chemical Abstracts, Beilstein, Patent databases, Dictionary of Natural Products, SetupX/BinBase, KNApSAcK DB, and finally those databases which were obtained by computational approaches, i.e. RiceCyc, KEGG, and Reactome. More than 5,000 small molecules were retrieved when searching these databases. Unfortunately, most often, genuine rice metabolites were retrieved together with non-metabolite database entries such as pesticides. Overlaps from database compound lists were very difficult to compare because structures were either not encoded in machine-readable format or because compound identifiers were not cross-referenced between databases.
Conclusions: We conclude that present databases are not capable of comprehensively retrieving all known metabolites. Metabolome lists are yet mostly restricted to genome-reconstructed pathways. We suggest that providers of (bio)chemical databases enrich their database identifiers to PubChem IDs and InChIKeys to enable cross-database queries. In addition, peer-reviewed journal repositories need to mandate submission of structures and spectra in machine readable format to allow automated semantic annotation of articles containing chemical structures. Such changes in publication standards and database architectures will enable researchers to compile current knowledge about the metabolome of species, which may extend to derived information such as spectral libraries, organ-specific metabolites, and cross-study comparisons.
Conflict of interest statement
Figures
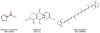
References
-
- International Rice Research Institute (IRRI); [ http://www.irri.org/]
-
- Kind T, Fiehn O. Hardware and Software Challenges for the Near Future: Structure Elucidation Concepts via Hyphenated Chromatographic Techniques. Lc Gc N Am. 2008;26:176.
-
- Hill DW, Kertesz TM, Fontaine D, Friedman R, Grant DF. Mass Spectral Metabonomics beyond Elemental Formula: Chemical Database Querying by Matching Experimental with Computational Fragmentation Spectra. Analytical Chemistry. 2008;80:5574–5582. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

