Determination of cellular lipids bound to human CD1d molecules
- PMID: 19415116
- PMCID: PMC2673035
- DOI: 10.1371/journal.pone.0005325
Determination of cellular lipids bound to human CD1d molecules
Abstract
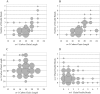
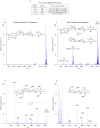
CD1 molecules are glycoproteins that present lipid antigens at the cell surface for immunological recognition by specialized populations of T lymphocytes. Prior experimental data suggest a wide variety of lipid species can bind to CD1 molecules, but little is known about the characteristics of cellular ligands that are selected for presentation. Here we have molecularly characterized lipids bound to the human CD1d isoform. Ligands were eluted from secreted CD1d molecules and separated by normal phase HPLC, then characterized by mass spectroscopy. A total of 177 lipid species were molecularly identified, comprising glycerophospholipids and sphingolipids. The glycerophospholipids included common diacylglycerol species, reduced forms known as plasmalogens, lyso-phospholipids (monoacyl species), and cardiolipins (tetraacyl species). The sphingolipids included sphingomyelins and glycosylated forms, such as the ganglioside GM3. These results demonstrate that human CD1d molecules bind a surprising diversity of lipid structures within the secretory pathway, including compounds that have been reported to play roles in cancer, autoimmune diseases, lipid signaling, and cell death.
Conflict of interest statement
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

