Transcriptional analysis of fracture healing and the induction of embryonic stem cell-related genes
- PMID: 19415118
- PMCID: PMC2673045
- DOI: 10.1371/journal.pone.0005393
Transcriptional analysis of fracture healing and the induction of embryonic stem cell-related genes
Abstract
Fractures are among the most common human traumas. Fracture healing represents a unique temporarily definable post-natal process in which to study the complex interactions of multiple molecular events that regulate endochondral skeletal tissue formation. Because of the regenerative nature of fracture healing, it is hypothesized that large numbers of post-natal stem cells are recruited and contribute to formation of the multiple cell lineages that contribute to this process. Bayesian modeling was used to generate the temporal profiles of the transcriptome during fracture healing. The temporal relationships between ontologies that are associated with various biologic, metabolic, and regulatory pathways were identified and related to developmental processes associated with skeletogenesis, vasculogenesis, and neurogenesis. The complement of all the expressed BMPs, Wnts, FGFs, and their receptors were related to the subsets of transcription factors that were concurrently expressed during fracture healing. We further defined during fracture healing the temporal patterns of expression for 174 of the 193 genes known to be associated with human genetic skeletal disorders. In order to identify the common regulatory features that might be present in stem cells that are recruited during fracture healing to other types of stem cells, we queried the transcriptome of fracture healing against that seen in embryonic stem cells (ESCs) and mesenchymal stem cells (MSCs). Approximately 300 known genes that are preferentially expressed in ESCs and approximately 350 of the known genes that are preferentially expressed in MSCs showed induction during fracture healing. Nanog, one of the central epigenetic regulators associated with ESC stem cell maintenance, was shown to be associated in multiple forms or bone repair as well as MSC differentiation. In summary, these data present the first temporal analysis of the transcriptome of an endochondral bone formation process that takes place during fracture healing. They show that neurogenesis as well as vasculogenesis are predominant components of skeletal tissue formation and suggest common pathways are shared between post-natal stem cells and those seen in ESCs.
Conflict of interest statement
Figures

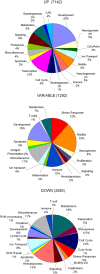

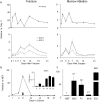
References
-
- US Department of Health and Human Services. 2004. Bone Health and Osteoporosis: A Report of the Surgeon General. Rockville: USDHHS/PHS. - PubMed
-
- Phillips AM. Overview of the fracture healing cascade. Injury. 2005;36S:55–57. - PubMed
-
- Nork SE. Chapter 47: Fractures of the shaft of the femur. In: Bucholz RW, Heckman JD, Court-Brown C, editors. Rockwood and Green's Fractures in Adults (6th edition) Lippincott, Williams, and Wilkins; 2006. pp. 1845–1914.
-
- Buckwalter JA, Einhorn TA, Marsh LJ. Bone and Joint Healing. In: Bucholz RW, Heckman JD, editors. Rockwood and Green's Fractures in Adults. Philadelphia: Lippincott, Williams, and Wilkins; 2001. pp. 245–271.
-
- Ferguson C, Alpern E, Miclau T, Helms JA. Does adult fracture repair recapitulate embryonic skeletal formation? Mech Dev. 1999;87:57–66. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

