Imaging transcription in living cells
- PMID: 19416065
- PMCID: PMC3166783
- DOI: 10.1146/annurev.biophys.050708.133728
Imaging transcription in living cells
Abstract
The advent of new technologies for the imaging of living cells has made it possible to determine the properties of transcription, the kinetics of polymerase movement, the association of transcription factors, and the progression of the polymerase on the gene. We report here the current state of the field and the progress necessary to achieve a more complete understanding of the various steps in transcription. Our Consortium is dedicated to developing and implementing the technology to further this understanding.
Figures
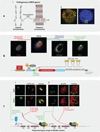
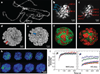
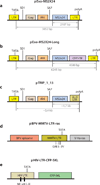
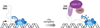
References
-
- Abramowitz M, Stegun IA. Handbook of Mathematical Functions. New York: Dover; 1965.
-
- Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat. Rev. Genet. 2007;8:507–517. - PubMed
-
- Beach DL, Salmon ED, Bloom K. Localization and anchoring of mRNA in budding yeast. Curr. Biol. 1999;9:569–578. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

