Single-molecule approaches to stochastic gene expression
- PMID: 19416069
- PMCID: PMC3126657
- DOI: 10.1146/annurev.biophys.37.032807.125928
Single-molecule approaches to stochastic gene expression
Abstract
Both the transcription of mRNAs from genes and their subsequent translation into proteins are inherently stochastic biochemical events, and this randomness can lead to substantial cell-to-cell variability in mRNA and protein numbers in otherwise identical cells. Recently, a number of studies have greatly enhanced our understanding of stochastic processes in gene expression by utilizing new methods capable of counting individual mRNAs and proteins in cells. In this review, we examine the insights that these studies have yielded in the field of stochastic gene expression. In particular, we discuss how these studies have played in understanding the properties of bursts in gene expression. We also compare the array of different methods that have arisen for single mRNA and protein detection, highlighting their relative strengths and weaknesses. In conclusion, we point out further areas where single-molecule techniques applied to gene expression may lead to new discoveries.
Figures
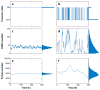
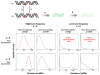
References
-
- Acar M, Becskei A, van Oudenaarden A. Enhancement of cellular memory by reducing stochastic transitions. Nature. 2005;435:228–32. - PubMed
-
- Acar M, Mettetal JT, van Oudenaarden A. Stochastic switching as a survival strategy in fluctuating environments. Nat Genet. 2008;40:471–75. - PubMed
-
- Bar-Even A, Paulsson J, Maheshri N, Carmi M, O’Shea E, et al. Noise in protein expression scales with natural protein abundance. Nat Genet. 2006;38:636–43. - PubMed
-
- Beach DL, Salmon ED, Bloom K. Localization and anchoring of mRNA in budding yeast. Curr Biol. 1999;9:569–78. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

