Peptidoglycan induces loss of a nuclear peptidoglycan recognition protein during host tissue development in a beneficial animal-bacterial symbiosis
- PMID: 19416268
- PMCID: PMC2758052
- DOI: 10.1111/j.1462-5822.2009.01315.x
Peptidoglycan induces loss of a nuclear peptidoglycan recognition protein during host tissue development in a beneficial animal-bacterial symbiosis
Abstract
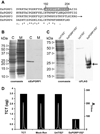
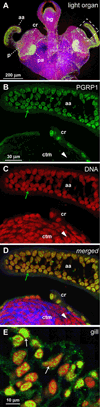
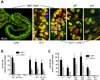
Peptidoglycan recognition proteins (PGRPs) are mediators of innate immunity and recently have been implicated in developmental regulation. To explore the interplay between these two roles, we characterized a PGRP in the host squid Euprymna scolopes (EsPGRP1) during colonization by the mutualistic bacterium Vibrio fischeri. Previous research on the squid-vibrio symbiosis had shown that, upon colonization of deep epithelium-lined crypts of the host light organ, symbiont-derived peptidoglycan monomers induce apoptosis-mediated regression of remote epithelial fields involved in the inoculation process. In this study, immunofluorescence microscopy revealed that EsPGRP1 localizes to the nuclei of epithelial cells, and symbiont colonization induces the loss of EsPGRP1 from apoptotic nuclei. The loss of nuclear EsPGRP1 occurred prior to DNA cleavage and breakdown of the nuclear membrane, but followed chromatin condensation, suggesting that it occurs during late-stage apoptosis. Experiments with purified peptidoglycan monomers and with V. fischeri mutants defective in peptidoglycan-monomer release provided evidence that these molecules trigger nuclear loss of EsPGRP1 and apoptosis. The demonstration of a nuclear PGRP is unprecedented, and the dynamics of EsPGRP1 during apoptosis provide a striking example of a connection between microbial recognition and developmental responses in the establishment of symbiosis.
Figures


Similar articles
-
Modulation of symbiont lipid A signaling by host alkaline phosphatases in the squid-vibrio symbiosis.mBio. 2012 May 1;3(3):e00093-12. doi: 10.1128/mBio.00093-12. Print 2012. mBio. 2012. PMID: 22550038 Free PMC article.
-
Recognition between symbiotic Vibrio fischeri and the haemocytes of Euprymna scolopes.Environ Microbiol. 2009 Feb;11(2):483-93. doi: 10.1111/j.1462-2920.2008.01788.x. Environ Microbiol. 2009. PMID: 19196278 Free PMC article.
-
Persistent symbiont colonization leads to a maturation of hemocyte response in the Euprymna scolopes/Vibrio fischeri symbiosis.Microbiologyopen. 2019 Oct;8(10):e858. doi: 10.1002/mbo3.858. Epub 2019 Jun 13. Microbiologyopen. 2019. PMID: 31197972 Free PMC article.
-
Host/microbe interactions revealed through "omics" in the symbiosis between the Hawaiian bobtail squid Euprymna scolopes and the bioluminescent bacterium Vibrio fischeri.Biol Bull. 2012 Aug;223(1):103-11. doi: 10.1086/BBLv223n1p103. Biol Bull. 2012. PMID: 22983036 Review.
-
The secret languages of coevolved symbioses: insights from the Euprymna scolopes-Vibrio fischeri symbiosis.Semin Immunol. 2012 Feb;24(1):3-8. doi: 10.1016/j.smim.2011.11.006. Epub 2011 Dec 7. Semin Immunol. 2012. PMID: 22154556 Free PMC article. Review.
Cited by
-
Use of Hybridization Chain Reaction-Fluorescent In Situ Hybridization To Track Gene Expression by Both Partners during Initiation of Symbiosis.Appl Environ Microbiol. 2015 Jul;81(14):4728-35. doi: 10.1128/AEM.00890-15. Epub 2015 May 8. Appl Environ Microbiol. 2015. PMID: 25956763 Free PMC article.
-
A peptidoglycan-recognition protein orchestrates the first steps of symbiont recruitment in the squid-vibrio symbiosis.Symbiosis. 2022 May;87(1):31-43. doi: 10.1007/s13199-022-00855-y. Epub 2022 Aug 6. Symbiosis. 2022. PMID: 36177150 Free PMC article.
-
Influence of Wolbachia on host gene expression in an obligatory symbiosis.BMC Microbiol. 2012 Jan 18;12 Suppl 1(Suppl 1):S7. doi: 10.1186/1471-2180-12-S1-S7. BMC Microbiol. 2012. PMID: 22376153 Free PMC article.
-
A Small Molecule Coordinates Symbiotic Behaviors in a Host Organ.mBio. 2021 Mar 9;12(2):e03637-20. doi: 10.1128/mBio.03637-20. mBio. 2021. PMID: 33688014 Free PMC article.
-
Differential expression of immune receptors in two marine sponges upon exposure to microbial-associated molecular patterns.Sci Rep. 2018 Oct 31;8(1):16081. doi: 10.1038/s41598-018-34330-w. Sci Rep. 2018. PMID: 30382170 Free PMC article.
References
-
- Apicella MA, Griffiss JM, Schneider H. Isolation and characterization of lipopolysaccharides, lipooligosaccharides, and lipid A. Methods Enzymol. 1994;235:242–252. - PubMed
-
- Beg AA, Baltimore D. An essential role for NF-κB in preventing TNF-alpha -induced cell death. Science. 1996;274:782–784. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

