Xylose reductase from Pichia stipitis with altered coenzyme preference improves ethanolic xylose fermentation by recombinant Saccharomyces cerevisiae
- PMID: 19416504
- PMCID: PMC2688486
- DOI: 10.1186/1754-6834-2-9
Xylose reductase from Pichia stipitis with altered coenzyme preference improves ethanolic xylose fermentation by recombinant Saccharomyces cerevisiae
Abstract
Background: Xylose reductase (XR) and xylitol dehydrogenase (XDH) from Pichia stipitis are the two enzymes most commonly used in recombinant Saccharomyces cerevisiae strains engineered for xylose utilization. The availability of NAD+ for XDH is limited during anaerobic xylose fermentation because of the preference of XR for NADPH. This in turn results in xylitol formation and reduced ethanol yield. The coenzyme preference of P. stipitis XR was changed by site-directed mutagenesis with the aim to engineer it towards NADH-preference.
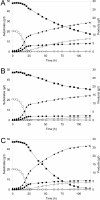
Results: XR variants were evaluated in S. cerevisiae strains with the following genetic modifications: overexpressed native P. stipitis XDH, overexpressed xylulokinase, overexpressed non-oxidative pentose phosphate pathway and deleted GRE3 gene encoding an NADPH dependent aldose reductase. All overexpressed genes were chromosomally integrated to ensure stable expression. Crude extracts of four different strains overexpressing genes encoding native P. stipitis XR, K270M and K270R mutants, as well as Candida parapsilosis XR, were enzymatically characterized. The physiological effects of the mutations were investigated in anaerobic xylose fermentation. The strain overexpressing P. stipitis XR with the K270R mutation gave an ethanol yield of 0.39 g (g consumed sugars)-1, a xylitol yield of 0.05 g (g consumed xylose)-1 and a xylose consumption rate of 0.28 g (g biomass)-1 h-1 in continuous fermentation at a dilution rate of 0.12 h-1, with 10 g l-1 glucose and 10 g l-1 xylose as carbon sources.
Conclusion: The cofactor preference of P. stipitis XR was altered by site-directed mutagenesis. When the K270R XR was combined with a metabolic engineering strategy that ensures high xylose utilization capabilities, a recombinant S. cerevisiae strain was created that provides a unique combination of high xylose consumption rate, high ethanol yield and low xylitol yield during ethanolic xylose fermentation.
Figures
References
-
- Hahn-Hägerdal B, Karhumaa K, Jeppsson M, Gorwa-Grauslund MF. Metabolic engineering for pentose utilization in Saccharomyces cerevisiae. Adv Biochem Eng Biotechnol. 2007;108:147–177. - PubMed
-
- van Maris AJ, Winkler AA, Kuyper M, de Laat WT, van Dijken JP, Pronk JT. Development of efficient xylose fermentation in Saccharomyces cerevisiae: xylose isomerase as a key component. Adv Biochem Eng Biotechnol. 2007;108:179–204. - PubMed
-
- Chu BC, Lee H. Genetic improvement of Saccharomyces cerevisiae for xylose fermentation. Biotechnol Adv. 2007;25:425–441. - PubMed
-
- Kötter P, Ciriacy M. Xylose fermentation by Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 1993;38:776–783.
-
- Tantirungkij M, Nakashima N, Seki T, Yoshida T. Construction of xylose-assimilating Saccharomyces cerevisiae. J Ferm Bioeng. 1993;75:83–88.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

