Catalytically-active complex of HIV-1 integrase with a viral DNA substrate binds anti-integrase drugs
- PMID: 19416821
- PMCID: PMC2688900
- DOI: 10.1073/pnas.0811919106
Catalytically-active complex of HIV-1 integrase with a viral DNA substrate binds anti-integrase drugs
Abstract
HIV-1 integration into the host cell genome is a multistep process catalyzed by the virally-encoded integrase (IN) protein. In view of the difficulty of obtaining a stable DNA-bound IN at high concentration as required for structure determination, we selected IN-DNA complexes that form disulfide linkages between 5'-thiolated DNA and several single mutations to cysteine around the catalytic site of IN. Mild reducing conditions allowed for selection of the most thermodynamically-stable disulfide-linked species. The most stable complexes induce tetramer formation of IN, as happens during the physiological integration reaction, and are able to catalyze the strand transfer step of retroviral integration. One of these complexes also binds strand-transfer inhibitors of HIV antiviral drugs, making it uniquely valuable among the mutants of this set for understanding portions of the integration reaction. This novel complex may help define substrate interactions and delineate the mechanism of action of known integration inhibitors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

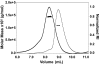



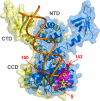
References
-
- Kukolj G, Skalka AM. Enhanced and coordinated processing of synapsed viral DNA ends by retroviral integrases in vitro. Genes Dev. 1995;9:2556–2567. - PubMed
-
- Brown PO. In: Retroviruses. Coffin JM, Hughes SH, Varmus HE, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1997. pp. 161–203. - PubMed
-
- Deprez E, et al. Oligomeric states of the HIV-1 integrase as measured by time-resolved fluorescence anisotropy. Biochemistry. 2000;39:9275–9284. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

