7SK snRNP/P-TEFb couples transcription elongation with alternative splicing and is essential for vertebrate development
- PMID: 19416841
- PMCID: PMC2683122
- DOI: 10.1073/pnas.0903188106
7SK snRNP/P-TEFb couples transcription elongation with alternative splicing and is essential for vertebrate development
Abstract
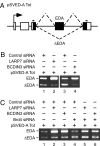
Eukaryotic gene expression is commonly controlled at the level of RNA polymerase II (RNAPII) pausing subsequent to transcription initiation. Transcription elongation is stimulated by the positive transcription elongation factor b (P-TEFb) kinase, which is suppressed within the 7SK small nuclear ribonucleoprotein (7SK snRNP). However, the biogenesis and functional significance of 7SK snRNP remain poorly understood. Here, we report that LARP7, BCDIN3, and the noncoding 7SK small nuclear RNA (7SK) are vital for the formation and stability of a cell stress-resistant core 7SK snRNP. Our functional studies demonstrate that 7SK snRNP is not only critical for controlling transcription elongation, but also for regulating alternative splicing of pre-mRNAs. Using a transient expression splicing assay, we find that 7SK snRNP disintegration promotes inclusion of an alternative exon via the increased occupancy of P-TEFb, Ser2-phosphorylated (Ser2-P) RNAPII, and the splicing factor SF2/ASF at the minigene. Importantly, knockdown of larp7 or bcdin3 orthologues in zebrafish embryos destabilizes 7SK and causes severe developmental defects and aberrant splicing of analyzed transcripts. These findings reveal a key role for P-TEFb in coupling transcription elongation with alternative splicing, and suggest that maintaining core 7SK snRNP is essential for vertebrate development.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Ni Z, Schwartz BE, Werner J, Suarez JR, Lis JT. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol Cell. 2004;13:55–65. - PubMed
-
- Ahn SH, Kim M, Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol Cell. 2004;13:67–76. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

