Haplotype structure strongly affects recombination in a maize genetic interval polymorphic for Helitron and retrotransposon insertions
- PMID: 19416860
- PMCID: PMC2688972
- DOI: 10.1073/pnas.0902972106
Haplotype structure strongly affects recombination in a maize genetic interval polymorphic for Helitron and retrotransposon insertions
Abstract
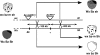
We have asked here how the remarkable variation in maize haplotype structure affects recombination. We compared recombination across a genetic interval of 9S in 2 highly dissimilar heterozygotes that shared 1 parent. The genetic interval in the common haplotype is approximately 100 kb long and contains 6 genes interspersed with gene-fragment-bearing Helitrons and retrotransposons that, together, comprise 70% of its length. In one heterozygote, most intergenic insertions are homozygous, although polymorphic, enabling us to determine whether any recombination junctions fall within them. In the other, most intergenic insertions are hemizygous and, thus, incapable of homologous recombination. Our analysis of the frequency and distribution of recombination in the interval revealed that: (i) Most junctions were circumscribed to the gene space, where they showed a highly nonuniform distribution. In both heterozygotes, more than half of the junctions fell in the stc1 gene, making it a clear recombination hotspot in the region. However, the genetic size of stc1 was 2-fold lower when flanked by a hemizygous 25-kb retrotransposon cluster. (ii) No junctions fell in the hypro1 gene in either heterozygote, making it a genic recombination coldspot. (iii) No recombination occurred within the gene fragments borne on Helitrons nor within retrotransposons, so neither insertion class contributes to the interval's genetic length. (iv) Unexpectedly, several junctions fell in an intergenic region not shared by all 3 haplotypes. (v) In general, the ability of a sequence to recombine correlated inversely with its methylation status. Our results show that haplotypic structural variability strongly affects the frequency and distribution of recombination events in maize.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Dooner HK, et al. A molecular genetic analysis of insertion mutations in the bronze locus in maize. Mol Gen Genet. 1985;200:240–246.
-
- Dooner HK, Hsia A-P, Schnable PS. In: Handbook of Maize: Domestication, Genetics, and Genome. Bennetzen JL, Hake SC, editors. New York: Springer Science; 2009. pp. 377–403.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

