Quantitative genome-scale analysis of protein localization in an asymmetric bacterium
- PMID: 19416866
- PMCID: PMC2671984
- DOI: 10.1073/pnas.0901781106
Quantitative genome-scale analysis of protein localization in an asymmetric bacterium
Abstract
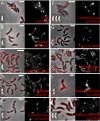
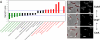
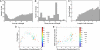
Despite the importance of subcellular localization for cellular activities, the lack of high-throughput, high-resolution imaging and quantitation methodologies has limited genomic localization analysis to a small number of archival studies focused on C-terminal fluorescent protein fusions. Here, we develop a high-throughput pipeline for generating, imaging, and quantitating fluorescent protein fusions that we use for the quantitative genomic assessment of the distributions of both N- and C-terminal fluorescent protein fusions. We identify nearly 300 localized Caulobacter crescentus proteins, up to 10-fold more than were previously characterized. The localized proteins tend to be involved in spatially or temporally dynamic processes and proteins that function together and often localize together as well. The distributions of the localized proteins were quantitated by using our recently described projected system of internal coordinates from interpolated contours (PSICIC) image analysis toolkit, leading to the identification of cellular regions that are over- or under-enriched in localized proteins and of potential differences in the mechanisms that target proteins to different subcellular destinations. The Caulobacter localizome data thus represent a resource for studying both global properties of protein localization and specific protein functions, whereas the localization analysis pipeline is a methodological resource that can be readily applied to other systems.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Huh WK, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. - PubMed
-
- Kitagawa M, et al. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): Unique resources for biological research. DNA Res. 2005;12:291–299. - PubMed
-
- Matsuyama A, et al. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2006;24:841–847. - PubMed
-
- Gitai Z. The new bacterial cell biology: Moving parts and subcellular architecture. Cell. 2005;120:577–586. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

