Experimental characterization of disordered and ordered aggregates populated during the process of amyloid fibril formation
- PMID: 19416886
- PMCID: PMC2683116
- DOI: 10.1073/pnas.0812227106
Experimental characterization of disordered and ordered aggregates populated during the process of amyloid fibril formation
Abstract
Recent experimental evidence points to intermediates populated during the process of amyloid fibril formation as the toxic moieties primarily responsible for the development of increasingly common disorders such as Alzheimer's disease and type II diabetes. We describe here the application of a pulse-labeling hydrogen-deuterium (HD) exchange strategy monitored by mass spectrometry (MS) and NMR spectroscopy (NMR) to characterize the aggregation process of an SH3 domain under 2 different conditions, both of which ultimately lead to well-defined amyloid fibrils. Under one condition, the intermediates appear to be largely amorphous in nature, whereas under the other condition protofibrillar species are clearly evident. Under the conditions favoring amorphous-like intermediates, only species having no protection against HD exchange can be detected in addition to the mature fibrils that show a high degree of protection. By contrast, under the conditions favoring protofibrillar-like intermediates, MS reveals that multiple species are present with different degrees of HD exchange protection, indicating that aggregation occurs initially through relatively disordered species that subsequently evolve to form ordered aggregates that eventually lead to amyloid fibrils. Further analysis using NMR provides residue-specific information on the structural reorganizations that take place during aggregation, as well as on the time scales by which they occur.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

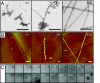

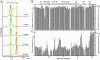
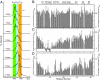
References
-
- Selkoe DJ. Folding proteins in fatal ways. Nature. 2003;426:900–904. - PubMed
-
- Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. - PubMed
-
- Rochet JC, Lansbury PT., Jr Amyloid fibrillogenesis: Themes and variations. Curr Opin Struct Biol. 2000;10:60–68. - PubMed
-
- Cleary JP, et al. Natural oligomers of the amyloid-beta protein specifically disrupt cognitive function. Nat Neurosci. 2005;8:79–84. - PubMed
-
- Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: Lessons from the Alzheimer's amyloid beta-peptide. Nat Rev Mol Cell Biol. 2007;8:101–112. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

