Niche partitioning in the coevolution of 2 distinct RNA enzymes
- PMID: 19416904
- PMCID: PMC2683078
- DOI: 10.1073/pnas.0903397106
Niche partitioning in the coevolution of 2 distinct RNA enzymes
Abstract
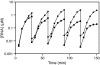
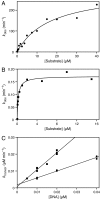
Organisms that compete for limited resources within a common environment may evolve traits that allow them to exploit distinct ecological niches, thus enabling multiple species to coexist within the same habitat. The process of niche partitioning now has been captured at the molecular level, employing the method of continuous in vitro evolution. Mixed populations of 2 different "species" of RNA enzymes were made to compete for limited amounts of one or more substrates, with utilization of the substrate being necessary for amplification of the RNA. Evolution in the presence of a single substrate led to the extinction of one or the other enzyme, whereas evolution in the presence of 5 alternative substrates led to the accumulation of mutations that allowed each enzyme to exploit a different preferred resource. The evolved enzymes were capable of sustained coevolution within a common environment, exemplifying the emergence of stable ecological niche behavior in a model system. Biochemical characterization of the 2 evolved enzymes revealed marked differences in their kinetic properties and adaptive strategies. One enzyme reacted with its preferred substrate approximately 100-fold faster than the other, but the slower-reacting species produced 2- to 3-fold more progeny per reacted parent molecule. The in vitro coevolution of 2 or more species of RNA enzymes will make possible further studies in molecular ecology, including the exploration of more complex behaviors, such as predation or cooperation, under controlled laboratory conditions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Bennett AF, Dao KM, Lenski RE. Rapid evolution in response to high-temperature selection. Nature. 1990;346:79–81. - PubMed
-
- Elena SF, Cooper VS, Lenski RE. Punctuated evolution caused by selection of rare beneficial mutations. Science. 1996;272:1802–1804. - PubMed
-
- Hillis DM, Bull JJ, White ME, Badgett MR, Molineux IJ. Experimental phylogenetics: Generation of a known phylogeny. Science. 1992;255:589–592. - PubMed
-
- Johns GC, Joyce GF. The promise and peril of continuous in vitro evolution. J Mol Evol. 2005;61:253–263. - PubMed
-
- Wright MC, Joyce GF. Continuous in vitro evolution of catalytic function. Science. 1997;276:614–617. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

