Functional pre- mRNA trans-splicing of coactivator CoAA and corepressor RBM4 during stem/progenitor cell differentiation
- PMID: 19416963
- PMCID: PMC2709364
- DOI: 10.1074/jbc.M109.006999
Functional pre- mRNA trans-splicing of coactivator CoAA and corepressor RBM4 during stem/progenitor cell differentiation
Abstract
Alternative splicing yields functionally distinctive gene products, and their balance plays critical roles in cell differentiation and development. We have previously shown that tumor-associated enhancer loss in coactivator gene CoAA leads to its altered alternative splicing. Here we identified two intergenic splicing variants, a zinc finger-containing coactivator CoAZ and a non-coding transcript ncCoAZ, between CoAA and its downstream corepressor gene RBM4. During stem/progenitor cell neural differentiation, we found that the switched alternative splicing and trans-splicing between CoAA and RBM4 transcripts result in lineage-specific expression of wild type CoAA, RBM4, and their variants. Stable expression of CoAA, RBM4, or their variants prevents the switch and disrupts the embryoid body formation. In addition, CoAA and RBM4 counter-regulate the target gene Tau at exon 10, and their splicing activities are subjected to the control by each splice variant. Further phylogenetic analysis showed that mammalian CoAA and RBM4 genes share common ancestry with the Drosophila melanogaster gene Lark, which is known to regulate early development and circadian rhythms. Thus, the trans-splicing between CoAA and RBM4 transcripts may represent a required regulation preserved during evolution. Our results demonstrate that a linked splicing control of transcriptional coactivator and corepressor is involved in stem/progenitor cell differentiation. The alternative splicing imbalance of CoAA and RBM4, because of loss of their common enhancer in cancer, may deregulate stem/progenitor cell differentiation.
Figures




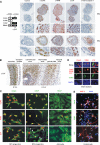

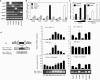
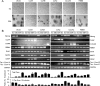
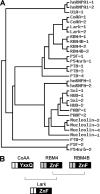
References
-
- Chow L. T., Gelinas R. E., Broker T. R., Roberts R. J. ( 1977) Cell 12, 1– 8 - PubMed
-
- Modrek B., Lee C. ( 2002) Nat. Genet. 30, 13– 19 - PubMed
-
- Sanford J. R., Caceres J. F. ( 2004) J. Cell Sci. 117, 6261– 6263 - PubMed
-
- Pan Q., Shai O., Lee L. J., Frey B. J., Blencowe B. J. ( 2008) Nat. Genet. 40, 1413– 1415 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

