The C terminus of Na+,K+-ATPase controls Na+ affinity on both sides of the membrane through Arg935
- PMID: 19416970
- PMCID: PMC2707196
- DOI: 10.1074/jbc.M109.015099
The C terminus of Na+,K+-ATPase controls Na+ affinity on both sides of the membrane through Arg935
Abstract
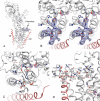
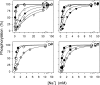
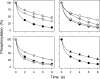
The Na(+),K(+)-ATPase C terminus has a unique location between transmembrane segments, appearing to participate in a network of interactions. We have examined the functional consequences of amino acid substitutions in this region and deletions of the C terminus of varying lengths. Assays revealing separately the mutational effects on internally and externally facing Na(+) sites, as well as E(1)-E(2) conformational changes, have been applied. The results pinpoint the two terminal tyrosines, Tyr(1017) and Tyr(1018), as well as putative interaction partners, Arg(935) in the loop between transmembrane segments M8 and M9 and Lys(768) in transmembrane segment M5, as crucial to Na(+) activation of phosphorylation of E(1), a partial reaction reflecting Na(+) interaction on the cytoplasmic side of the membrane. Tyr(1017), Tyr(1018), and Arg(935) are furthermore indispensable to Na(+) interaction on the extracellular side of the membrane, as revealed by inability of high Na(+) concentrations to drive the transition from E(1)P to E(2)P backwards toward E(1)P and inhibit Na(+)-ATPase activity in mutants. Lys(768) is not important for Na(+) binding from the external side of the membrane but is involved in stabilization of the E(2) form. These data demonstrate that the C terminus controls Na(+) affinity on both sides of the membrane and suggest that Arg(935) constitutes an important link between the C terminus and the third Na(+) site, involving an arginine-pi stacking interaction between Arg(935) and the C-terminal tyrosines. Lys(768) may interact preferentially with the C terminus in E(1) and E(1)P forms and with the loop between transmembrane segments M6 and M7 in E(2) and E(2)P forms.
Figures
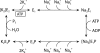


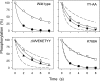

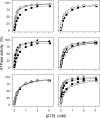
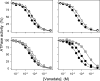
References
-
- Kaplan J. H. (2002) Annu. Rev. Biochem. 71, 511–535 - PubMed
-
- Post R. L., Hegyvary C., Kume S. (1972) J. Biol. Chem. 247, 6530–6540 - PubMed
-
- Morth J. P., Pedersen B. P., Toustrup-Jensen M. S., Sørensen T. L., Petersen J., Andersen J. P., Vilsen B., Nissen P. (2007) Nature 450, 1043–1049 - PubMed
-
- Toyoshima C., Nakasako M., Nomura H., Ogawa H. (2000) Nature 405, 647–655 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

