Genome-wide replication profiles indicate an expansive role for Rpd3L in regulating replication initiation timing or efficiency, and reveal genomic loci of Rpd3 function in Saccharomyces cerevisiae
- PMID: 19417103
- PMCID: PMC2682954
- DOI: 10.1101/gad.1784309
Genome-wide replication profiles indicate an expansive role for Rpd3L in regulating replication initiation timing or efficiency, and reveal genomic loci of Rpd3 function in Saccharomyces cerevisiae
Abstract
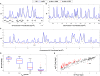
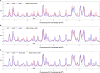
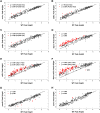
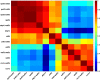
In higher eukaryotes, heritable gene silencing is associated with histone deacetylation and late replication timing. In Saccharomyces cerevisiae, the histone deacetylase Rpd3 regulates gene expression and also modulates replication timing; however, these mechanisms have been suggested to be independent, and no global association has been found between replication timing and gene expression levels. Using 5-Bromo-2'-deoxyuridine (BrdU) incorporation to generate genome-wide replication profiles, we identified >100 late-firing replication origins that are regulated by Rpd3L, which is specifically targeted to promoters to silence transcription. Rpd3S, which recompacts chromatin after transcription, plays a primary role at only a handful of origins, but subtly influences initiation timing globally. The ability of these functionally distinct Rpd3 complexes to affect replication initiation timing supports the idea that histone deacetylation directly influences initiation timing. Accordingly, loss of Rpd3 function results in higher levels of histone H3 and H4 acetylation surrounding Rpd3-regulated origins, and these origins show a significant association with Rpd3 chromatin binding and gene regulation, supporting a general link between histone acetylation, replication timing, and control of gene expression in budding yeast. Our results also reveal a novel and complementary genomic map of Rpd3L- and Rpd3S-regulated chromosomal loci.
Figures
Similar articles
-
The Rpd3-Sin3 histone deacetylase regulates replication timing and enables intra-S origin control in Saccharomyces cerevisiae.Mol Cell Biol. 2004 Jun;24(11):4769-80. doi: 10.1128/MCB.24.11.4769-4780.2004. Mol Cell Biol. 2004. PMID: 15143171 Free PMC article.
-
Rpd3 regulates single-copy origins independently of the rDNA array by opposing Fkh1-mediated origin stimulation.Proc Natl Acad Sci U S A. 2022 Oct 4;119(40):e2212134119. doi: 10.1073/pnas.2212134119. Epub 2022 Sep 26. Proc Natl Acad Sci U S A. 2022. PMID: 36161938 Free PMC article.
-
Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription.Cell. 2005 Nov 18;123(4):581-92. doi: 10.1016/j.cell.2005.10.023. Cell. 2005. PMID: 16286007
-
Genome-wide patterns of histone modifications in yeast.Nat Rev Mol Cell Biol. 2006 Sep;7(9):657-66. doi: 10.1038/nrm1986. Epub 2006 Aug 16. Nat Rev Mol Cell Biol. 2006. PMID: 16912715 Review.
-
A site to remember: H3K36 methylation a mark for histone deacetylation.Mutat Res. 2007 May 1;618(1-2):130-4. doi: 10.1016/j.mrfmmm.2006.08.014. Epub 2007 Jan 21. Mutat Res. 2007. PMID: 17346757 Review.
Cited by
-
Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading.Genome Res. 2010 Feb;20(2):201-11. doi: 10.1101/gr.097873.109. Epub 2009 Dec 7. Genome Res. 2010. PMID: 19996087 Free PMC article.
-
Protein phosphatase 1 recruitment by Rif1 regulates DNA replication origin firing by counteracting DDK activity.Cell Rep. 2014 Apr 10;7(1):53-61. doi: 10.1016/j.celrep.2014.02.019. Epub 2014 Mar 20. Cell Rep. 2014. PMID: 24656819 Free PMC article.
-
Drosophila Mcm10 is required for DNA replication and differentiation in the compound eye.PLoS One. 2014 Mar 31;9(3):e93450. doi: 10.1371/journal.pone.0093450. eCollection 2014. PLoS One. 2014. PMID: 24686397 Free PMC article.
-
Origin Firing Regulations to Control Genome Replication Timing.Genes (Basel). 2019 Mar 6;10(3):199. doi: 10.3390/genes10030199. Genes (Basel). 2019. PMID: 30845782 Free PMC article. Review.
-
Inhibition of histone deacetylase in cancer cells slows down replication forks, activates dormant origins, and induces DNA damage.Cancer Res. 2010 Jun 1;70(11):4470-80. doi: 10.1158/0008-5472.CAN-09-3028. Epub 2010 May 11. Cancer Res. 2010. PMID: 20460513 Free PMC article.
References
-
- Aggarwal B.D., Calvi B.R. Chromatin regulates origin activity in Drosophila follicle cells. Nature. 2004;430:372–376. - PubMed
-
- Aparicio O.M., Weinstein D.M., Bell S.P. Components and dynamics of DNA replication complexes in S. cerevisiae: Redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. - PubMed
-
- Bell S.P., Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
