Complexity in transcription control at the activation domain-mediator interface
- PMID: 19417216
- PMCID: PMC2774526
- DOI: 10.1126/scisignal.1164302
Complexity in transcription control at the activation domain-mediator interface
Erratum in
- Sci Signal. 2009 Sep 22;2(89):er8
Abstract
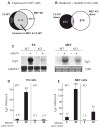
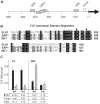
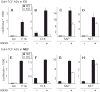
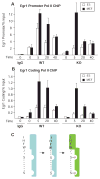
Transcript elongation by polymerase II paused at the Egr1 promoter is activated by mitogen-activated protein kinase phosphorylation of the ternary complex factor (TCF) ELK1 bound at multiple upstream sites and subsequent phospho-ELK1 interaction with mediator through the MED23 subunit. Consequently, Med23 knockout (KO) nearly eliminates Egr1 (early growth response factor 1) transcription in embryonic stem (ES) cells, leaving a paused polymerase at the promoter. Med23 KO did not, however, eliminate Egr1 transcription in fibroblasts. Chromatin immunoprecipitation analysis and direct visualization of fluorescently labeled TCF derivatives and mediator subunits revealed that three closely related TCFs bound to the same control regions. The relative amounts of these TCFs, which responded differently to the loss of MED23, differed in ES cells and fibroblasts. Transcriptome analysis suggests that most genes expressed in both cell types, such as Egr1, are regulated by alternative transcription factors in the two cell types that respond differently to the same signal transduction pathways.
Figures
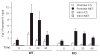
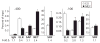

References
-
- Drysdale CM, Jackson BM, McVeigh R, Klebanow ER, Bai Y, Kokubo T, Swanson M, Nakatani Y, Weil PA, Hinnebusch AG. The Gcn4p activation domain interacts specifically in vitro with RNA polymerase II holoenzyme, TFIID, and the Adap-Gcn5p coactivator complex. Mol Cell Biol. 1998;18:1711–1724. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

