Single molecule analysis of a red fluorescent RecA protein reveals a defect in nucleoprotein filament nucleation that relates to its reduced biological functions
- PMID: 19419960
- PMCID: PMC2707236
- DOI: 10.1074/jbc.M109.004895
Single molecule analysis of a red fluorescent RecA protein reveals a defect in nucleoprotein filament nucleation that relates to its reduced biological functions
Abstract
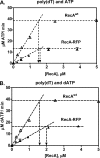
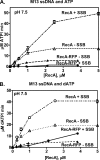
Fluorescent fusion proteins are exceedingly useful for monitoring protein localization in situ or visualizing protein behavior at the single molecule level. Unfortunately, some proteins are rendered inactive by the fusion. To circumvent this problem, we fused a hyperactive RecA protein (RecA803 protein) to monomeric red fluorescent protein (mRFP1) to produce a functional protein (RecA-RFP) that is suitable for in vivo and in vitro analysis. In vivo, the RecA-RFP partially restores UV resistance, conjugational recombination, and SOS induction to recA(-) cells. In vitro, the purified RecA-RFP protein forms a nucleoprotein filament whose k(cat) for single-stranded DNA-dependent ATPase activity is reduced approximately 3-fold relative to wild-type protein, and which is largely inhibited by single-stranded DNA-binding protein. However, RecA protein is also a dATPase; dATP supports RecA-RFP nucleoprotein filament formation in the presence of single-stranded DNA-binding protein. Furthermore, as for the wild-type protein, the activities of RecA-RFP are further enhanced by shifting the pH to 6.2. As a consequence, RecA-RFP is proficient for DNA strand exchange with dATP or at lower pH. Finally, using single molecule visualization, RecA-RFP was seen to assemble into a continuous filament on duplex DNA, and to extend the DNA approximately 1.7-fold. Consistent with its attenuated activities, RecA-RFP nucleates onto double-stranded DNA approximately 3-fold more slowly than the wild-type protein, but still requires approximately 3 monomers to form the rate-limited nucleus needed for filament assembly. Thus, RecA-RFP reveals that its attenuated biological functions correlate with a reduced frequency of nucleoprotein filament nucleation at the single molecule level.
Figures



References
-
- Chudakov D. M., Lukyanov S., Lukyanov K. A. (2005) Trends Biotechnol. 23, 605–613 - PubMed
-
- Nie S., Chiu D. T., Zare R. N. (1994) Science 266, 1018–1021 - PubMed
-
- Funatsu T., Harada Y., Tokunaga M., Saito K., Yanagida T. (1995) Nature 374, 555–559 - PubMed
-
- Noji H., Yasuda R., Yoshida M., Kinosita K., Jr. (1997) Nature 386, 299–302 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

