ProSMoS server: a pattern-based search using interaction matrix representation of protein structures
- PMID: 19420061
- PMCID: PMC2703969
- DOI: 10.1093/nar/gkp316
ProSMoS server: a pattern-based search using interaction matrix representation of protein structures
Abstract
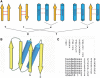
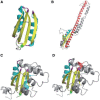
Assessing structural similarity and defining common regions through comparison of protein spatial structures is an important task in functional and evolutionary studies of proteins. There are many servers that compare structures and define sub-structures in common between proteins through superposition and closeness of either coordinates or contacts. However, a natural way to analyze a structure for experts working on structure classification is to look for specific three-dimensional (3D) motifs and patterns instead of finding common features in two proteins. Such motifs can be described by the architecture and topology of major secondary structural elements (SSEs) without consideration of subtle differences in 3D coordinates. Despite the importance of motif-based structure searches, currently there is a shortage of servers to perform this task. Widely known TOPS does not fully address this problem, as it finds only topological match but does not take into account other important spatial properties, such as interactions and chirality. Here, we implemented our approach to protein structure pattern search (ProSMoS) as a web-server. ProSMoS converts 3D structure into an interaction matrix representation including the SSE types, handednesses of connections between SSEs, coordinates of SSE starts and ends, types of interactions between SSEs and beta-sheet definitions. For a user-defined structure pattern, ProSMoS lists all structures from a database that contain this pattern. ProSMoS server will be of interest to structural biologists who would like to analyze very general and distant structural similarities. The ProSMoS web server is available at: http://prodata.swmed.edu/ProSMoS/.
Figures
Similar articles
-
deconSTRUCT: general purpose protein database search on the substructure level.Nucleic Acids Res. 2010 Jul;38(Web Server issue):W590-4. doi: 10.1093/nar/gkq489. Epub 2010 Jun 3. Nucleic Acids Res. 2010. PMID: 20522512 Free PMC article.
-
Searching for three-dimensional secondary structural patterns in proteins with ProSMoS.Bioinformatics. 2007 Jun 1;23(11):1331-8. doi: 10.1093/bioinformatics/btm121. Epub 2007 Mar 24. Bioinformatics. 2007. PMID: 17384423
-
HorA web server to infer homology between proteins using sequence and structural similarity.Nucleic Acids Res. 2009 Jul;37(Web Server issue):W532-8. doi: 10.1093/nar/gkp328. Epub 2009 May 5. Nucleic Acids Res. 2009. PMID: 19417074 Free PMC article.
-
QSalignWeb: A Server to Predict and Analyze Protein Quaternary Structure.Front Mol Biosci. 2022 Jan 5;8:787510. doi: 10.3389/fmolb.2021.787510. eCollection 2021. Front Mol Biosci. 2022. PMID: 35071324 Free PMC article. Review.
-
Protein structure databases.Mol Biotechnol. 2011 Jun;48(2):183-98. doi: 10.1007/s12033-010-9372-4. Mol Biotechnol. 2011. PMID: 21225378 Review.
Cited by
-
Fast and accurate protein substructure searching with simulated annealing and GPUs.BMC Bioinformatics. 2010 Sep 3;11:446. doi: 10.1186/1471-2105-11-446. BMC Bioinformatics. 2010. PMID: 20813068 Free PMC article.
-
iPBA: a tool for protein structure comparison using sequence alignment strategies.Nucleic Acids Res. 2011 Jul;39(Web Server issue):W18-23. doi: 10.1093/nar/gkr333. Epub 2011 May 17. Nucleic Acids Res. 2011. PMID: 21586582 Free PMC article.
-
deconSTRUCT: general purpose protein database search on the substructure level.Nucleic Acids Res. 2010 Jul;38(Web Server issue):W590-4. doi: 10.1093/nar/gkq489. Epub 2010 Jun 3. Nucleic Acids Res. 2010. PMID: 20522512 Free PMC article.
-
CASP9 target classification.Proteins. 2011;79 Suppl 10(Suppl 10):21-36. doi: 10.1002/prot.23190. Epub 2011 Oct 14. Proteins. 2011. PMID: 21997778 Free PMC article.
-
BCSearch: fast structural fragment mining over large collections of protein structures.Nucleic Acids Res. 2015 Jul 1;43(W1):W378-82. doi: 10.1093/nar/gkv492. Epub 2015 May 14. Nucleic Acids Res. 2015. PMID: 25977292 Free PMC article.
References
-
- Anantharaman V, Aravind L, Koonin EV. Emergence of diverse biochemical activities in evolutionarily conserved structural scaffolds of proteins. Curr. Opin. Chem. Biol. 2003;7:12–20. - PubMed
-
- Ammelburg M, Hartmann MD, Djuranovic S, Alva V, Koretke KK, Martin J, Sauer G, Truffault V, Zeth K, Lupas AN, et al. A CTP-dependent archaeal riboflavin kinase forms a bridge in the evolution of cradle-loop barrels. Structure. 2007;15:1577–1590. - PubMed
-
- Grishin NV. Mh1 domain of Smad is a degraded homing endonuclease. J. Mol. Biol. 2001;307:31–37. - PubMed
-
- Koehl P. Protein structure similarities. Curr. Opin. Struct. Biol. 2001;11:348–353. - PubMed
-
- Eidhammer I, Jonassen I, Taylor WR. Structure comparison and structure patterns. J. Comput. Biol. 2000;7:685–716. - PubMed

