Requirement for XLF/Cernunnos in alignment-based gap filling by DNA polymerases lambda and mu for nonhomologous end joining in human whole-cell extracts
- PMID: 19420065
- PMCID: PMC2709571
- DOI: 10.1093/nar/gkp283
Requirement for XLF/Cernunnos in alignment-based gap filling by DNA polymerases lambda and mu for nonhomologous end joining in human whole-cell extracts
Abstract
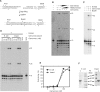
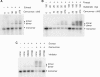
XLF/Cernunnos is a core protein of the nonhomologous end-joining pathway of DNA double-strand break repair. To better define the role of Cernunnos in end joining, whole-cell extracts were prepared from Cernunnos-deficient human cells. These extracts effected little joining of DNA ends with cohesive 5' or 3' overhangs, and no joining at all of partially complementary 3' overhangs that required gap filling prior to ligation. Assays in which gap-filled but unligated intermediates were trapped using dideoxynucleotides revealed that there was no gap filling on aligned DSB ends in the Cernunnos-deficient extracts. Recombinant Cernunnos protein restored gap filling and end joining of partially complementary overhangs, and stimulated joining of cohesive ends more than twentyfold. XLF-dependent gap filling was nearly eliminated by immunodepletion of DNA polymerase lambda, but was restored by addition of either polymerase lambda or polymerase mu. Thus, Cernunnos is essential for gap filling by either polymerase during nonhomologous end joining, suggesting that it plays a major role in aligning the two DNA ends in the repair complex.
Figures


References
-
- Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124:301–313. - PubMed
-
- Buck D, Malivert L, de Chasseval R, Barraud A, Fondaneche MC, Sanal O, Plebani A, Stephan JL, Hufnagel M, Le Deist F, et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. - PubMed
-
- Andres SN, Modesti M, Tsai CJ, Chu G, Junop MS. Crystal structure of human XLF: a twist in nonhomologous DNA end-joining. Mol. Cell. 2007;28:1093–1101. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

