DASMI: exchanging, annotating and assessing molecular interaction data
- PMID: 19420069
- PMCID: PMC2677739
- DOI: 10.1093/bioinformatics/btp142
DASMI: exchanging, annotating and assessing molecular interaction data
Abstract
Motivation: Ever increasing amounts of biological interaction data are being accumulated worldwide, but they are currently not readily accessible to the biologist at a single site. New techniques are required for retrieving, sharing and presenting data spread over the Internet.
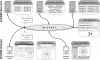
Results: We introduce the DASMI system for the dynamic exchange, annotation and assessment of molecular interaction data. DASMI is based on the widely used Distributed Annotation System (DAS) and consists of a data exchange specification, web servers for providing the interaction data and clients for data integration and visualization. The decentralized architecture of DASMI affords the online retrieval of the most recent data from distributed sources and databases. DASMI can also be extended easily by adding new data sources and clients. We describe all DASMI components and demonstrate their use for protein and domain interactions.
Availability: The DASMI tools are available at http://www.dasmi.de/ and http://ipfam.sanger.ac.uk/graph. The DAS registry and the DAS 1.53E specification is found at http://www.dasregistry.org/.
Figures



References
-
- Aloy P, Russell RB. Structural systems biology: modelling protein interactions. Nat. Rev. Mol. Cell Biol. 2006;7:188–197. - PubMed
-
- Aragues R, et al. PIANA: protein interactions and network analysis. Bioinformatics. 2006;22:1015–1017. - PubMed
-
- Avila-Campillo I, et al. BioNetBuilder: automatic integration of biological networks. Bioinformatics. 2007;23:392–393. - PubMed

