A high speed detection platform based on surface-enhanced Raman scattering for monitoring antibiotic-induced chemical changes in bacteria cell wall
- PMID: 19421405
- PMCID: PMC2674953
- DOI: 10.1371/journal.pone.0005470
A high speed detection platform based on surface-enhanced Raman scattering for monitoring antibiotic-induced chemical changes in bacteria cell wall
Abstract
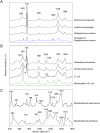
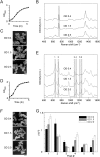
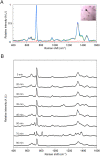
Rapid and accurate diagnosis for pathogens and their antibiotic susceptibility is critical for controlling bacterial infections. Conventional methods for determining bacterium's sensitivity to antibiotic depend mostly on measuring the change of microbial proliferation in response to the drug. Such "biological assay" inevitably takes time, ranging from days for fast-growing bacteria to weeks for slow-growers. Here, a novel tool has been developed to detect the "chemical features" of bacterial cell wall that enables rapid identification of drug resistant bacteria within hours. The surface-enhanced Raman scattering (SERS) technique based on our newly developed SERS-active substrate was applied to assess the fine structures of the bacterial cell wall. The SERS profiles recorded by such a platform are sensitive and stable, that could readily reflect different bacterial cell walls found in Gram-positive, Gram-negative, or mycobacteria groups. Moreover, characteristic changes in SERS profile were noticed in the drug-sensitive bacteria at the early period (i.e., approximately 1 hr) of antibiotic exposure, which could be used to differentiate them from the drug-resistant ones. The SERS-based diagnosis could be applied to a single bacterium. The high-speed SERS detection represents a novel approach for microbial diagnostics. The single-bacterium detection capability of SERS makes possible analyses directly on clinical specimen instead of pure cultured bacteria.
Conflict of interest statement
Figures


Similar articles
-
Antibiotic Susceptibility Test with Surface-Enhanced Raman Scattering in a Microfluidic System.Anal Chem. 2019 Sep 3;91(17):10988-10995. doi: 10.1021/acs.analchem.9b01027. Epub 2019 Aug 19. Anal Chem. 2019. PMID: 31387345
-
Surface-enhanced Raman scattering for the rapid discrimination of bacteria.Faraday Discuss. 2006;132:281-92; discussion 309-19. doi: 10.1039/b506413a. Faraday Discuss. 2006. PMID: 16833123
-
Rapid single-cell detection and identification of pathogens by using surface-enhanced Raman spectroscopy.Analyst. 2017 May 21;142(10):1782-1789. doi: 10.1039/c7an00106a. Epub 2017 Apr 21. Analyst. 2017. PMID: 28430277
-
Surface-Enhanced Raman Scattering for Rapid Detection and Characterization of Antibiotic-Resistant Bacteria.Adv Healthc Mater. 2018 Jul;7(13):e1701335. doi: 10.1002/adhm.201701335. Epub 2018 Mar 5. Adv Healthc Mater. 2018. PMID: 29504273 Review.
-
Recent Progress of Surface-Enhanced Raman Spectroscopy for Bacteria Detection.Biosensors (Basel). 2023 Mar 6;13(3):350. doi: 10.3390/bios13030350. Biosensors (Basel). 2023. PMID: 36979564 Free PMC article. Review.
Cited by
-
The biochemical origins of the surface-enhanced Raman spectra of bacteria: a metabolomics profiling by SERS.Anal Bioanal Chem. 2016 Jul;408(17):4631-47. doi: 10.1007/s00216-016-9540-x. Epub 2016 Apr 21. Anal Bioanal Chem. 2016. PMID: 27100230 Free PMC article.
-
RNA markers enable phenotypic test of antibiotic susceptibility in Neisseria gonorrhoeae after 10 minutes of ciprofloxacin exposure.Sci Rep. 2018 Aug 2;8(1):11606. doi: 10.1038/s41598-018-29707-w. Sci Rep. 2018. PMID: 30072794 Free PMC article.
-
Recent Advances in Bacterial Detection Using Surface-Enhanced Raman Scattering.Biosensors (Basel). 2024 Aug 1;14(8):375. doi: 10.3390/bios14080375. Biosensors (Basel). 2024. PMID: 39194603 Free PMC article. Review.
-
Antibiotic Susceptibility Testing with Raman Biosensing.Antibiotics (Basel). 2022 Dec 14;11(12):1812. doi: 10.3390/antibiotics11121812. Antibiotics (Basel). 2022. PMID: 36551469 Free PMC article. Review.
-
Asymmetric split-ring plasmonic nanostructures for the optical sensing of Escherichia coli.Biomed Opt Express. 2023 Aug 25;14(9):4875-4887. doi: 10.1364/BOE.497820. eCollection 2023 Sep 1. Biomed Opt Express. 2023. PMID: 37791281 Free PMC article.
References
-
- Mothershed EA, Whitney AM. Nucleic acid-based methods for the detection of bacterial pathogens: present and future considerations for the clinical laboratory. Clin Chim Acta. 2006;363:206–220. - PubMed
-
- Rolain JM, Mallet MN, Fournier PE, Raoult D. Real-time PCR for universal antibiotic susceptibility testing. J Antimicrob Chemother. 2004;54:538–541. - PubMed
-
- Kneip K, Moskovits M, Kneip H. Surface-Enhanced Raman Scattering: Physics and Applications Topics in Applied Physics. 2006;103:1–464.
-
- Kennedy BJSS, Dickey M, Carron KT. Determination of the distance dependence and experimental effects for modified SERS substrates based on self-assembled monolayers fourmed using alkanethiols. J Phys Chem B. 1999;103:3640–3646.
-
- Nie S, Emory SR. Probing Single Molecules and Single Nanoparticles by Surface-Enhanced Raman Scattering. Science. 1997;275:1102–1106. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

