Genomics, transcriptional profiling, and heart failure
- PMID: 19422981
- PMCID: PMC2738978
- DOI: 10.1016/j.jacc.2008.12.064
Genomics, transcriptional profiling, and heart failure
Abstract
Associated with technological progress in deoxyribonucleic acid and messenger ribonucleic acid profiling, advances in basic biology have led to a more complete and sophisticated understanding of interactions among genes, environment, and affected tissues in the setting of complex and heterogeneous conditions such as heart failure (HF). Ongoing identification of mutations causing hereditary hypertrophic and dilated cardiomyopathies has provided both pathophysiological insights and clinically applicable diagnostics for these relatively rare conditions. Genotyping clinical trial participants and genome-wide association studies have accelerated the identification of much more common disease- and treatment-modifying genes that explain patient-to-patient differences that have long been recognized by practicing clinicians. At the same time, increasingly detailed characterization of gene expression within diseased tissues and circulating cells from animal models and patients are providing new insights into the pathophysiology of HF that permit identification of novel diagnostic and therapeutic targets. In this rapidly evolving field, there is already ample support for the concept that genetic and expression profiling can enhance diagnostic sensitivity and specificity while providing a rational basis for prioritizing alternative therapeutic options for patients with cardiomyopathies and HF. Although the extensive characterizations provided by genomic and transcriptional profiling will increasingly challenge clinicians' abilities to utilize complex and diverse information, advances in clinical information technology and user interfaces will permit greater individualization of prevention and treatment strategies to address the HF epidemic.
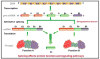
Figures
References
-
- Guttmacher AE, Collins FS. Genomic medicine--a primer. N Engl J Med. 2002;347:1512–20. - PubMed
-
- Khoury MJ. Genetics and genomics in practice: the continuum from genetic disease to genetic information in health and disease. Genet Med. 2003;5:261–8. - PubMed
-
- Alcalai R, Seidman JG, Seidman CE. Genetic basis of hypertrophic cardiomyopathy: from bench to the clinics. J Cardiovasc Electrophysiol. 2008;19:104–10. - PubMed
-
- Bowles KR, Bowles NE. Genetics of inherited cardiomyopathies. Expert Rev Cardiovasc Ther. 2004;2:683–97. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

