Population structure of Streptococcus oralis
- PMID: 19423627
- PMCID: PMC2885674
- DOI: 10.1099/mic.0.027284-0
Population structure of Streptococcus oralis
Abstract
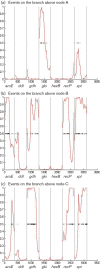
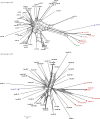
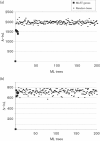
Streptococcus oralis is a member of the normal human oral microbiota, capable of opportunistic pathogenicity; like related oral streptococci, it exhibits appreciable phenotypic and genetic variation. A multilocus sequence typing (MLST) scheme for S. oralis was developed and the resultant data analysed to examine the population structure of the species. Analysis of 113 isolates, confirmed as belonging to the S. oralis/mitis group by 16S rRNA gene sequencing, characterized the population as highly diverse and undergoing inter- and intra-species recombination with a probable clonal complex structure. ClonalFrame analysis of these S. oralis isolates along with examples of Streptococcus pneumoniae, Streptococcus mitis and Streptococcus pseudopneumoniae grouped the named species into distinct, coherent populations and did not support the clustering of S. pseudopneumoniae with S. mitis as reported previously using distance-based methods. Analysis of the individual loci suggested that this discrepancy was due to the possible hybrid nature of S. pseudopneumoniae. The data are available on the public MLST website (http://pubmlst.org/soralis/).
Figures


References
-
- Arbique, J. C., Poyart, C., Trieu-Cuot, P., Quesne, G., Carvalho Mda, G., Steigerwalt, A. G., Morey, R. E., Jackson, D., Davidson, R. J. & Facklam, R. R. (2004). Accuracy of phenotypic and genotypic testing for identification of Streptococcus pneumoniae and description of Streptococcus pseudopneumoniae sp. nov. J Clin Microbiol 42, 4686–4696. - PMC - PubMed
-
- Bek-Thomsen, M., Tettelin, H., Hance, I., Nelson, K. E. & Kilian, M. (2008). Population diversity and dynamics of Streptococcus mitis, Streptococcus oralis, and Streptococcus infantis in the upper respiratory tracts of adults, determined by a nonculture strategy. Infect Immun 76, 1889–1896. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

