Neuroserpin polymers activate NF-kappaB by a calcium signaling pathway that is independent of the unfolded protein response
- PMID: 19423713
- PMCID: PMC2709363
- DOI: 10.1074/jbc.M109.010744
Neuroserpin polymers activate NF-kappaB by a calcium signaling pathway that is independent of the unfolded protein response
Abstract
The autosomal dominant dementia familial encephalopathy with neuroserpin inclusion bodies is characterized by the accumulation of ordered polymers of mutant neuroserpin within the endoplasmic reticulum of neurones. We show here that intracellular neuroserpin polymers activate NF-kappaB by a pathway that is independent of the IRE1, ATF6, and PERK limbs of the canonical unfolded protein response but is dependent on intracellular calcium. This pathway provides a mechanism for cells to sense and react to the accumulation of folded structures of mutant serpins within the endoplasmic reticulum. Our results provide strong support for the endoplasmic reticulum overload response being independent of the unfolded protein response.
Figures
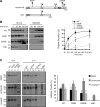


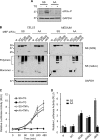
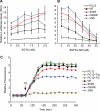
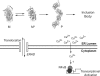
References
-
- Carrell R. W., Lomas D. A. ( 1997) Lancet 350, 134– 138 - PubMed
-
- Lomas D. A., Carrell R. W. ( 2002) Nat. Rev. Genet. 3, 759– 768 - PubMed
-
- Sivasothy P., Dafforn T. R., Gettins P. G., Lomas D. A. ( 2000) J. Biol. Chem. 275, 33663– 33668 - PubMed
-
- Davis R. L., Shrimpton A. E., Holohan P. D., Bradshaw C., Feiglin D., Collins G. H., Sonderegger P., Kinter J., Becker L. M., Lacbawan F., Krasnewich D., Muenke M., Lawrence D. A., Yerby M. S., Shaw C. M., Gooptu B., Elliott P. R., Finch J. T., Carrell R. W., Lomas D. A. ( 1999) Nature 401, 376– 379 - PubMed

