Chromatin remodelling beyond transcription: the INO80 and SWR1 complexes
- PMID: 19424290
- PMCID: PMC6103619
- DOI: 10.1038/nrm2693
Chromatin remodelling beyond transcription: the INO80 and SWR1 complexes
Abstract
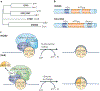
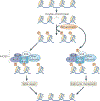
Chromatin-modifying factors have essential roles in DNA processing pathways that dictate cellular functions. The ability of chromatin modifiers, including the INO80 and SWR1 chromatin-remodelling complexes, to regulate transcriptional processes is well established. However, recent studies reveal that the INO80 and SWR1 complexes have crucial functions in many other essential processes, including DNA repair, checkpoint regulation, DNA replication, telomere maintenance and chromosome segregation. During these diverse nuclear processes, the INO80 and SWR1 complexes function cooperatively with their histone substrates, gamma-H2AX and H2AZ. This research reveals that INO80 and SWR1 ATP-dependent chromatin remodelling is an integral component of pathways that maintain genomic integrity.
Figures



References
-
- Gorbalenya AE & Koonin EV Helicases: amino acid sequence comparisons and structure–function relationships. Curr. Opin. Struct. Biol. 3, 419–429 (1993).
-
-
Shen X, Mizuguchi G, Hamiche A & Wu C A chromatin remodelling complex involved in transcription and DNA processing. Nature 406, 541–544 (2000).
Identifies and characterizes the yeast INO80 chromatin-remodelling complex, the founding member of the INO80 subfamily.
-
-
- Mizuguchi G et al. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303, 343–348 (2004). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

