HIV evolution in early infection: selection pressures, patterns of insertion and deletion, and the impact of APOBEC
- PMID: 19424423
- PMCID: PMC2671846
- DOI: 10.1371/journal.ppat.1000414
HIV evolution in early infection: selection pressures, patterns of insertion and deletion, and the impact of APOBEC
Abstract
The pattern of viral diversification in newly infected individuals provides information about the host environment and immune responses typically experienced by the newly transmitted virus. For example, sites that tend to evolve rapidly across multiple early-infection patients could be involved in enabling escape from common early immune responses, could represent adaptation for rapid growth in a newly infected host, or could represent reversion from less fit forms of the virus that were selected for immune escape in previous hosts. Here we investigated the diversification of HIV-1 env coding sequences in 81 very early B subtype infections previously shown to have resulted from transmission or expansion of single viruses (n = 78) or two closely related viruses (n = 3). In these cases, the sequence of the infecting virus can be estimated accurately, enabling inference of both the direction of substitutions as well as distinction between insertion and deletion events. By integrating information across multiple acutely infected hosts, we find evidence of adaptive evolution of HIV-1 env and identify a subset of codon sites that diversified more rapidly than can be explained by a model of neutral evolution. Of 24 such rapidly diversifying sites, 14 were either i) clustered and embedded in CTL epitopes that were verified experimentally or predicted based on the individual's HLA or ii) in a nucleotide context indicative of APOBEC-mediated G-to-A substitutions, despite having excluded heavily hypermutated sequences prior to the analysis. In several cases, a rapidly evolving site was embedded both in an APOBEC motif and in a CTL epitope, suggesting that APOBEC may facilitate early immune escape. Ten rapidly diversifying sites could not be explained by CTL escape or APOBEC hypermutation, including the most frequently mutated site, in the fusion peptide of gp41. We also examined the distribution, extent, and sequence context of insertions and deletions, and we provide evidence that the length variation seen in hypervariable loop regions of the envelope glycoprotein is a consequence of selection and not of mutational hotspots. Our results provide a detailed view of the process of diversification of HIV-1 following transmission, highlighting the role of CTL escape and hypermutation in shaping viral evolution during the establishment of new infections.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
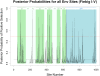



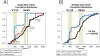
References
-
- Friedrich TC, Dodds EJ, Yant LJ, Vojnov L, Rudersdorf R, et al. Reversion of CTL escape-variant immunodeficiency viruses in vivo. Nature Medicine. 2004;10:275–281. - PubMed
-
- Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: Fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. - PubMed
-
- Blish CA, Nedellec R, Mandaliya K, Mosier DE, Overbaugh J. HIV-1 subtype A envelope variants from early in infection have variable sensitivity to neutralization and to inhibitors of viral entry. Aids. 2007;21:693–702. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

