Diurnally entrained anticipatory behavior in archaea
- PMID: 19424498
- PMCID: PMC2675056
- DOI: 10.1371/journal.pone.0005485
Diurnally entrained anticipatory behavior in archaea
Abstract
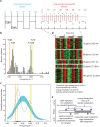
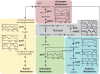
By sensing changes in one or few environmental factors biological systems can anticipate future changes in multiple factors over a wide range of time scales (daily to seasonal). This anticipatory behavior is important to the fitness of diverse species, and in context of the diurnal cycle it is overall typical of eukaryotes and some photoautotrophic bacteria but is yet to be observed in archaea. Here, we report the first observation of light-dark (LD)-entrained diurnal oscillatory transcription in up to 12% of all genes of a halophilic archaeon Halobacterium salinarum NRC-1. Significantly, the diurnally entrained transcription was observed under constant darkness after removal of the LD stimulus (free-running rhythms). The memory of diurnal entrainment was also associated with the synchronization of oxic and anoxic physiologies to the LD cycle. Our results suggest that under nutrient limited conditions halophilic archaea take advantage of the causal influence of sunlight (via temperature) on O(2) diffusivity in a closed hypersaline environment to streamline their physiology and operate oxically during nighttime and anoxically during daytime.
Conflict of interest statement
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

