Targeted deletion of the gamma-adducin gene (Add3) in mice reveals differences in alpha-adducin interactions in erythroid and nonerythroid cells
- PMID: 19425068
- PMCID: PMC2827150
- DOI: 10.1002/ajh.21427
Targeted deletion of the gamma-adducin gene (Add3) in mice reveals differences in alpha-adducin interactions in erythroid and nonerythroid cells
Abstract
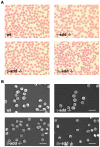
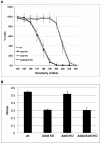
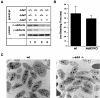
In red blood cells (RBCs) adducin heterotetramers localize to the spectrin-actin junction of the peripheral membrane skeleton. We previously reported that deletion of beta-adducin results in osmotically fragile, microcytic RBCs and a phenotype of hereditary spherocytosis (HS). Notably, alpha-adducin was significantly reduced, while gamma-adducin, normally present in limited amounts, was increased approximately 5-fold, suggesting that alpha-adducin requires a heterologous binding partner for stability and function, and that gamma-adducin can partially substitute for the absence of beta-adducin. To test these assumptions we generated gamma-adducin null mice. gamma-adducin null RBCs appear normal on Wright's stained peripheral blood smears and by scanning electron microscopy. All membrane skeleton proteins examined are present in normal amounts, and all hematological parameters measured are normal. Despite a loss of approximately 70% of alpha-adducin in gamma-adducin null platelets, no bleeding defect is observed and platelet structure appears normal. Moreover, systemic blood pressure and pulse are normal in gamma-adducin null mice. gamma- and beta-adducin null mice were intercrossed to generate double null mice. Loss of gamma-adducin does not exacerbate the beta-adducin null HS phenotype although the amount alpha-adducin is reduced to barely detectable levels. The stability of alpha-adducin in the absence of a heterologous binding partner varies considerably in various tissues. The amount of alpha-adducin is modestly reduced ( approximately 15%) in the kidney, while in the spleen and brain is reduced by approximately 50% with the loss of a heterologous beta- or gamma-adducin binding partner. These results suggest that the structural properties of adducin differ significantly between erythroid and various nonerythroid cell types.
Figures



References
-
- Suriyapperuma SP, Lozovatsky L, Ciciotte SL, Peters LL, Gilligan DM. The mouse adducin gene family: alternative splicing and chromosomal localization. Mamm Genome. 2000;11:16–23. - PubMed
-
- Fowler VM. Identification and purification of a novel Mr 43,000 tropomyosin-binding protein from human erythrocyte membranes. J Biol Chem. 1987;262:12792–12800. - PubMed
-
- Dong L, Chapline C, Mousseau B, et al. 35H, a sequence isolated as a protein kinase C binding protein, is a novel member of the adducin family. J Biol Chem. 1995;270:25534–25540. - PubMed
-
- Bennett V, Baines AJ. Spectrin and ankyrin-based pathways: metazoan inventions for integrating cells into tissues. Physiol Rev. 2001;81:1353–1392. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
