TGF-beta signaling proteins and the Protein Ontology
- PMID: 19426460
- PMCID: PMC2679403
- DOI: 10.1186/1471-2105-10-S5-S3
TGF-beta signaling proteins and the Protein Ontology
Abstract
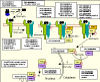
Background: The Protein Ontology (PRO) is designed as a formal and principled Open Biomedical Ontologies (OBO) Foundry ontology for proteins. The components of PRO extend from a classification of proteins on the basis of evolutionary relationships at the homeomorphic level to the representation of the multiple protein forms of a gene, including those resulting from alternative splicing, cleavage and/or post-translational modifications. Focusing specifically on the TGF-beta signaling proteins, we describe the building, curation, usage and dissemination of PRO.
Results: PRO is manually curated on the basis of PrePRO, an automatically generated file with content derived from standard protein data sources. Manual curation ensures that the treatment of the protein classes and the internal and external relationships conform to the PRO framework. The current release of PRO is based upon experimental data from mouse and human proteins wherein equivalent protein forms are represented by single terms. In addition to the PRO ontology, the annotation of PRO terms is released as a separate PRO association file, which contains, for each given PRO term, an annotation from the experimentally characterized sub-types as well as the corresponding database identifiers and sequence coordinates. The annotations are added in the form of relationship to other ontologies. Whenever possible, equivalent forms in other species are listed to facilitate cross-species comparison. Splice and allelic variants, gene fusion products and modified protein forms are all represented as entities in the ontology. Therefore, PRO provides for the representation of protein entities and a resource for describing the associated data. This makes PRO useful both for proteomics studies where isoforms and modified forms must be differentiated, and for studies of biological pathways, where representations need to take account of the different ways in which the cascade of events may depend on specific protein modifications.
Conclusion: PRO provides a framework for the formal representation of protein classes and protein forms in the OBO Foundry. It is designed to enable data retrieval and integration and machine reasoning at the molecular level of proteins, thereby facilitating cross-species comparisons, pathway analysis, disease modeling and the generation of new hypotheses.
Figures



References
-
- Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, OBI Consortium. Leontis N, Rocca-Serra P, Ruttenberg A, Sansone SA, Scheuermann RH, Shah N, Whetzel PL, Lewis S. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol. 2007;25:1251–1255. - PMC - PubMed
-
- Sidhu AS, Dillon TS, et al. Protein Ontology: Data Integration using Protein Ontology. In: Ma Z, Chen JY, editor. Database Modeling in Biology: Practices and Challenges. New York, Springer Inc; 2006. pp. 39–60.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

