Lrig1 expression defines a distinct multipotent stem cell population in mammalian epidermis
- PMID: 19427292
- PMCID: PMC2698066
- DOI: 10.1016/j.stem.2009.04.014
Lrig1 expression defines a distinct multipotent stem cell population in mammalian epidermis
Abstract
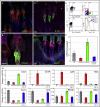
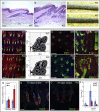
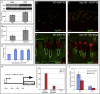
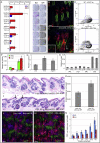
Lrig1 is a marker of human interfollicular epidermal stem cells and helps maintain stem cell quiescence. We show that, in mouse epidermis, Lrig1 defines the hair follicle junctional zone adjacent to the sebaceous glands and infundibulum. Lrig1 is a Myc target gene; loss of Lrig1 increases the proliferative capacity of stem cells in culture and results in epidermal hyperproliferation in vivo. Lrig1-expressing cells can give rise to all of the adult epidermal lineages in skin reconstitution assays. However, during homeostasis and on retinoic acid stimulation, they are bipotent, contributing to the sebaceous gland and interfollicular epidermis. beta-catenin activation increases the size of the junctional zone compartment, and loss of Lrig1 causes a selective increase in beta-catenin-induced ectopic hair follicle formation in the interfollicular epidermis. Our results suggest that Lrig1-positive cells constitute a previously unidentified reservoir of adult mouse interfollicular epidermal stem cells.
Figures



References
-
- Arnold I., Watt F.M. c-Myc activation in transgenic mouse epidermis results in mobilization of stem cells and differentiation of their progeny. Curr. Biol. 2001;11:558–568. - PubMed
-
- Blanpain C., Lowry W.E., Geoghegan A., Polak L., Fuchs E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118:635–648. - PubMed
-
- Bleul C.C., Corbeaux T., Reuter A., Fisch P., Monting J.S., Boehm T. Formation of a functional thymus initiated by a postnatal epithelial progenitor cell. Nature. 2006;441:992–996. - PubMed
-
- Braun K.M., Niemann C., Jensen U.B., Sundberg J.P., Silva-Vargas V., Watt F.M. Manipulation of stem cell proliferation and lineage commitment: Visualisation of label-retaining cells in wholemounts of mouse epidermis. Development. 2003;130:5241–5255. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

