Arylphosphonium salts interact with DNA to modulate cytotoxicity
- PMID: 19429515
- PMCID: PMC2712833
- DOI: 10.1016/j.mrgentox.2009.01.002
Arylphosphonium salts interact with DNA to modulate cytotoxicity
Erratum in
- Mutat Res. 2009 May 31;676(1-2):131
Abstract
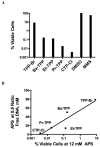
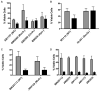
Arylphosphonium salts (APS) are compounds that have both lipophilic and cationic character, allowing them facile transport through plasma membranes or cell walls to accumulate in the cytoplasm or mitochondria of cells. APS molecules preferentially accumulate in tumor cells and are therefore under investigation as tumor imaging agents and mitochondrial targeting molecules. We have generated a systematic set of APS to study their ability to associate with DNA. The chemical structure of the APS determines the extent of its interaction with DNA and therefore its ability to aggregate the DNA. Also, APS compounds blocked DNA amplification in vitro at concentrations below the aggregation threshold, corroborating the structure/interaction relationship. Furthermore, the extent of APS:DNA interaction strongly correlates with bacterial toxicity, implying that APS molecules may deter cellular metabolic DNA pathways. Finally, DNA repair deficient and DNA bypass polymerase deficient bacterial strains were screened for sensitivity to APS. Interestingly, no single pathway for the repair or tolerance of these compounds was solely responsible for APS mediated toxicity. Taken together, these findings suggest that APS compounds may be capable of targeting and regulating unchecked cell growth and therefore show potential applications as a chemotherapeutic agent.
Figures



Similar articles
-
Anticancer activity of salinomycin quaternary phosphonium salts.Eur J Med Chem. 2025 Jan 15;282:117055. doi: 10.1016/j.ejmech.2024.117055. Epub 2024 Nov 14. Eur J Med Chem. 2025. PMID: 39556896
-
Synthesis and biological evaluation of arylphosphonium-benzoxaborole conjugates as novel anticancer agents.Bioorg Med Chem Lett. 2020 Jul 15;30(14):127259. doi: 10.1016/j.bmcl.2020.127259. Epub 2020 May 12. Bioorg Med Chem Lett. 2020. PMID: 32527557
-
The Study of Anti-/Pro-Oxidant, Lipophilic, Microbial and Spectroscopic Properties of New Alkali Metal Salts of 5-O-Caffeoylquinic Acid.Int J Mol Sci. 2018 Feb 4;19(2):463. doi: 10.3390/ijms19020463. Int J Mol Sci. 2018. PMID: 29401704 Free PMC article.
-
Antimicrobial polymeric materials with quaternary ammonium and phosphonium salts.Int J Mol Sci. 2015 Feb 6;16(2):3626-55. doi: 10.3390/ijms16023626. Int J Mol Sci. 2015. PMID: 25667977 Free PMC article. Review.
-
Activity of gemini quaternary ammonium salts against microorganisms.Appl Microbiol Biotechnol. 2019 Jan;103(2):625-632. doi: 10.1007/s00253-018-9523-2. Epub 2018 Nov 20. Appl Microbiol Biotechnol. 2019. PMID: 30460534 Review.
Cited by
-
The Remarkable and Selective In Vitro Cytotoxicity of Synthesized Bola-Amphiphilic Nanovesicles on Etoposide-Sensitive and -Resistant Neuroblastoma Cells.Nanomaterials (Basel). 2024 Sep 16;14(18):1505. doi: 10.3390/nano14181505. Nanomaterials (Basel). 2024. PMID: 39330662 Free PMC article.
-
Preclinical evaluation of novel triphenylphosphonium salts with broad-spectrum activity.PLoS One. 2010 Oct 4;5(10):e13131. doi: 10.1371/journal.pone.0013131. PLoS One. 2010. PMID: 20957228 Free PMC article.
-
Palladium(ii), platinum(ii), and silver(i) complexes with 3-acetylcoumarin benzoylhydrazone Schiff base: Synthesis, characterization, biomolecular interactions, cytotoxic activity, and computational studies.RSC Adv. 2024 Jun 18;14(27):19512-19527. doi: 10.1039/d4ra02738h. eCollection 2024 Jun 12. RSC Adv. 2024. PMID: 38895519 Free PMC article.
-
Strongly ROS-Correlated, Time-Dependent, and Selective Antiproliferative Effects of Synthesized Nano Vesicles on BRAF Mutant Melanoma Cells and Their Hyaluronic Acid-Based Hydrogel Formulation.Int J Mol Sci. 2024 Sep 19;25(18):10071. doi: 10.3390/ijms251810071. Int J Mol Sci. 2024. PMID: 39337557 Free PMC article.
-
Discovery of Di(het)arylmethane and Dibenzoxanthene Derivatives as Potential Anticancer Agents.Int J Mol Sci. 2024 Jun 18;25(12):6724. doi: 10.3390/ijms25126724. Int J Mol Sci. 2024. PMID: 38928428 Free PMC article.
References
-
- Hendry LB, Mahesh VB, Bransome ED, Jr, Ewing DE. Small molecule intercalation with double stranded DNA: implications for normal gene regulation and for predicting the biological efficacy and genotoxicity of drugs and other chemicals. Mutat Res. 2007;623:53–71. - PubMed
-
- Rideout DC, Calogeropoulou T, Jaworski JS, Dagnino R, Jr, McCarthy MR. Phosphonium salts exhibiting selective anti-carcinoma activity in vitro. Anticancer Drug Des. 1989;4:265–280. - PubMed
-
- Madar I, Anderson JH, Szabo Z, Scheffel U, Kao PF, Ravert HT, Dannals RF. Enhanced uptake of [11C]TPMP in canine brain tumor: a PET study. J Nucl Med. 1999;40:1180–1185. - PubMed
-
- Madar I, Ravert H, Nelkin B, Abro M, Pomper M, Dannals R, Frost JJ. Characterization of membrane potential-dependent uptake of the novel PET tracer 18F-fluorobenzyl triphenylphosphonium cation. Eur J Nucl Med Mol Imaging. 2007;34:2057–2065. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

