orf4 of the Bacillus cereus sigB gene cluster encodes a general stress-inducible Dps-like bacterioferritin
- PMID: 19429618
- PMCID: PMC2704712
- DOI: 10.1128/JB.00272-09
orf4 of the Bacillus cereus sigB gene cluster encodes a general stress-inducible Dps-like bacterioferritin
Abstract
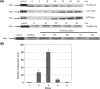
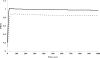
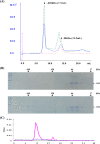
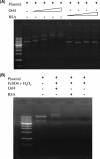
The function of orf4 in the sigB cluster in Bacillus cereus ATCC 14579 remains to be explored. Amino-acid sequence analysis has revealed that Orf4 is homologous with bacterioferritins and Dps. In this study, we generated an orf4-null mutant and produced recombinant protein rOrf4 to establish the role of orf4. In vitro, the purified rOrf4 was found to exist in two distinct forms, a dimeric form and a polymer form, through size exclusion analysis. The latter form exhibited a unique filament structure, in contrast to the typical spherical tetracosamer structure of bacterioferritins; the former can be induced to form rOrf4 polymers immediately after the addition of FeCl(2). Catalysis of the oxidation of ferrous irons by ferroxidase activity was detected with rOrf4, and the mineralized irons were subsequently sequestered only in the rOrf4 polymer. Moreover, rOrf4 exerted DNA-protective activity against oxidative damage via DNA binding in a nonspecific manner, as is seen with Dps. In vivo, deletion of orf4 had no effect on activation of the alternative sigma factor sigma(B), and therefore, orf4 is not associated with sigma(B) regulation; however, orf4 can be significantly upregulated upon environmental stress but not H(2)O(2) treatment. B. cereus strains with constitutive Orf4 expression exhibited a viability higher than that of the orf4-null mutant, under specific oxidative stress or heat shock. Taken together, these results suggest that Orf4 functions as a Dps-like bacterioferritin in response to environmental stress and can provide cell protection from oxidative damage through iron sequestration and DNA binding.
Figures



Similar articles
-
Bacterioferritin of Magnetospirillum gryphiswaldense Is a Heterotetraeicosameric Complex Composed of Functionally Distinct Subunits but Is Not Involved in Magnetite Biomineralization.mBio. 2019 May 21;10(3):e02795-18. doi: 10.1128/mBio.02795-18. mBio. 2019. PMID: 31113903 Free PMC article.
-
Desulfovibrio vulgaris bacterioferritin uses H(2)O(2) as a co-substrate for iron oxidation and reveals DPS-like DNA protection and binding activities.Biochem J. 2012 Aug 15;446(1):125-33. doi: 10.1042/BJ20111439. Biochem J. 2012. PMID: 22642556
-
Differential regulation and activity against oxidative stress of Dps proteins in Bacillus cereus.Int J Med Microbiol. 2013 Dec;303(8):662-73. doi: 10.1016/j.ijmm.2013.09.011. Int J Med Microbiol. 2013. PMID: 24383075
-
Ferritins, iron uptake and storage from the bacterioferritin viewpoint.EMBO J. 2003 May 1;22(9):1959-68. doi: 10.1093/emboj/cdg215. EMBO J. 2003. PMID: 12727864 Free PMC article. Review.
-
Iron storage in bacteria.Adv Microb Physiol. 1998;40:281-351. doi: 10.1016/s0065-2911(08)60134-4. Adv Microb Physiol. 1998. PMID: 9889981 Review.
Cited by
-
Bacillus safensis FO-36b and Bacillus pumilus SAFR-032: a whole genome comparison of two spacecraft assembly facility isolates.BMC Microbiol. 2018 Jun 8;18(1):57. doi: 10.1186/s12866-018-1191-y. BMC Microbiol. 2018. PMID: 29884123 Free PMC article.
-
Bacillus cereus cell response upon exposure to acid environment: toward the identification of potential biomarkers.Front Microbiol. 2013 Oct 2;4:284. doi: 10.3389/fmicb.2013.00284. Front Microbiol. 2013. PMID: 24106490 Free PMC article. Review.
-
SigB regulates stress resistance, glucose starvation, MnSOD production, biofilm formation, and root colonization in Bacillus cereus 905.Appl Microbiol Biotechnol. 2021 Aug;105(14-15):5943-5957. doi: 10.1007/s00253-021-11402-y. Epub 2021 Aug 5. Appl Microbiol Biotechnol. 2021. PMID: 34350477
-
Dps Is a Universally Conserved Dual-Action DNA-Binding and Ferritin Protein.J Bacteriol. 2022 May 17;204(5):e0003622. doi: 10.1128/jb.00036-22. Epub 2022 Apr 5. J Bacteriol. 2022. PMID: 35380871 Free PMC article. Review.
-
Structure and mechanism of iron translocation by a Dps protein from Microbacterium arborescens.J Biol Chem. 2011 Oct 7;286(40):34872-82. doi: 10.1074/jbc.M111.246108. Epub 2011 Jul 16. J Biol Chem. 2011. PMID: 21768097 Free PMC article.
References
-
- Aitken-Rogers, H., C. Singleton, A. Lewin, A. Taylor-Gee, G. R. Moore, and N. E. Le-Brun. 2004. Effect of phosphate on bacterioferritin-catalysed iron(II) oxidation. J. Biol. Inorg. Chem. 9161-170. - PubMed
-
- Almirón, M., A. J. Link, D. Furlong, and R. Kolter. 1992. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 62646-2654. - PubMed
-
- Andrews, S. C. 1998. Iron storage in bacteria. Adv. Microb. Physiol. 40281-351. - PubMed
-
- Andrews, S. C., N. E. L. Brun, V. Barynin, A. J. Thomson, G. R. Moore, J. R. Guest, and P. M. Harrison. 1995. Site-directed replacement of the coaxial heme ligands of bacterioferritin generates heme-free variants. J. Biol. Chem. 27023268-23274. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

