Interactions of DNA with biofilm-derived membrane vesicles
- PMID: 19429627
- PMCID: PMC2698485
- DOI: 10.1128/JB.00717-08
Interactions of DNA with biofilm-derived membrane vesicles
Abstract
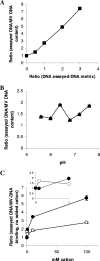
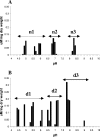
The biofilm matrix contributes to the chemistry, structure, and function of biofilms. Biofilm-derived membrane vesicles (MVs) and DNA, both matrix components, demonstrated concentration-, pH-, and cation-dependent interactions. Furthermore, MV-DNA association influenced MV surface properties. This bears consequences for the reactivity and availability for interaction of matrix polymers and other constituents.
Figures



References
-
- Allesen-Holm, M., K. B. Barken, L. Yang, M. Klausen, J. S. Webb, S. Kjelleberg, S. Molin, M. Givskov, and T. Tolker-Nielsen. 2006. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 591114-1128. - PubMed
-
- Beveridge, T. J., S. A. Makin, J. L. Kadurugamuwa, and Z. Li. 1997. Interactions between biofilms and the environment. FEMS Microbiol. Rev. 20291-303. - PubMed
-
- Böckelmann, U., A. Janke, R. Kuhn, T. R. Neu, J. Wecke, J. R. Lawrence, and U. Szewzyk. 2006. Bacterial extracellular DNA forming a defined network-like structure. FEMS Microbiol. Lett. 26231-38. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

