New insights into centromere organization and evolution from the white-cheeked gibbon and marmoset
- PMID: 19429672
- PMCID: PMC2734153
- DOI: 10.1093/molbev/msp101
New insights into centromere organization and evolution from the white-cheeked gibbon and marmoset
Abstract
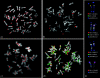
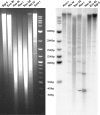
The evolutionary history of alpha-satellite DNA, the major component of primate centromeres, is hardly defined because of the difficulty in its sequence assembly and its rapid evolution when compared with most genomic sequences. By using several approaches, we have cloned, sequenced, and characterized alpha-satellite sequences from two species representing critical nodes in the primate phylogeny: the white-cheeked gibbon, a lesser ape, and marmoset, a New World monkey. Sequence analyses demonstrate that white-cheeked gibbon and marmoset alpha-satellite sequences are formed by units of approximately 171 and approximately 342 bp, respectively, and they both lack the high-order structure found in humans and great apes. Fluorescent in situ hybridization characterization shows a broad dispersal of alpha-satellite in the white-cheeked gibbon genome including centromeric, telomeric, and chromosomal interstitial localizations. On the other hand, centromeres in marmoset appear organized in highly divergent dimers roughly of 342 bp that show a similarity between monomers much lower than previously reported dimers, thus representing an ancient dimeric structure. All these data shed light on the evolution of the centromeric sequences in Primates. Our results suggest radical differences in the structure, organization, and evolution of alpha-satellite DNA among different primate species, supporting the notion that 1) all the centromeric sequence in Primates evolved by genomic amplification, unequal crossover, and sequence homogenization using a 171 bp monomer as the basic seeding unit and 2) centromeric function is linked to relatively short repeated elements, more than higher-order structure. Moreover, our data indicate that complex higher-order repeat structures are a peculiarity of the hominid lineage, showing the more complex organization in humans.
Figures



Similar articles
-
Comparative Analyses of Gibbon Centromeres Reveal Dynamic Genus-Specific Shifts in Repeat Composition.Mol Biol Evol. 2021 Aug 23;38(9):3972-3992. doi: 10.1093/molbev/msab148. Mol Biol Evol. 2021. PMID: 33983366 Free PMC article.
-
Repetitive sequences originating from the centromere constitute large-scale heterochromatin in the telomere region in the siamang, a small ape.Heredity (Edinb). 2012 Sep;109(3):180-7. doi: 10.1038/hdy.2012.28. Epub 2012 Jun 6. Heredity (Edinb). 2012. PMID: 22669075 Free PMC article.
-
Organization and molecular evolution of CENP-A--associated satellite DNA families in a basal primate genome.Genome Biol Evol. 2011;3:1136-49. doi: 10.1093/gbe/evr083. Epub 2011 Aug 9. Genome Biol Evol. 2011. PMID: 21828373 Free PMC article.
-
Dark Matter of Primate Genomes: Satellite DNA Repeats and Their Evolutionary Dynamics.Cells. 2020 Dec 18;9(12):2714. doi: 10.3390/cells9122714. Cells. 2020. PMID: 33352976 Free PMC article. Review.
-
Structural and functional dynamics of human centromeric chromatin.Annu Rev Genomics Hum Genet. 2006;7:301-13. doi: 10.1146/annurev.genom.7.080505.115613. Annu Rev Genomics Hum Genet. 2006. PMID: 16756479 Review.
Cited by
-
The evolutionary life cycle of the resilient centromere.Chromosoma. 2012 Aug;121(4):327-40. doi: 10.1007/s00412-012-0369-6. Epub 2012 Apr 11. Chromosoma. 2012. PMID: 22527114 Review.
-
Two types of alpha satellite DNA in distinct chromosomal locations in Azara's owl monkey.DNA Res. 2013 Jun;20(3):235-40. doi: 10.1093/dnares/dst004. Epub 2013 Mar 10. DNA Res. 2013. PMID: 23477842 Free PMC article.
-
Identification and characterization of a subtelomeric satellite DNA in Callitrichini monkeys.DNA Res. 2017 Aug 1;24(4):377-385. doi: 10.1093/dnares/dsx010. DNA Res. 2017. PMID: 28854689 Free PMC article.
-
Centromere remodeling in Hoolock leuconedys (Hylobatidae) by a new transposable element unique to the gibbons.Genome Biol Evol. 2012;4(7):648-58. doi: 10.1093/gbe/evs048. Epub 2012 May 16. Genome Biol Evol. 2012. PMID: 22593550 Free PMC article.
-
Locational diversity of alpha satellite DNA and intergeneric hybridization aspects in the Nomascus and Hylobates genera of small apes.PLoS One. 2014 Oct 7;9(10):e109151. doi: 10.1371/journal.pone.0109151. eCollection 2014. PLoS One. 2014. PMID: 25290445 Free PMC article.
References
-
- Alexandrov I, Kazakov A, Tumeneva I, Shepelev V, Yurov Y. Alpha-satellite DNA of primates: old and new families. Chromosoma. 2001;110:253–266. - PubMed
-
- Alexandrov IA, Mashkova TD, Romanova LY, Yurov YB, Kisselev LL. Segment substitutions in alpha satellite DNA. Unusual structure of human chromosome 3-specific alpha satellite repeat unit. J Mol Biol. 1993a;231:516–520. - PubMed
-
- Alexandrov IA, Mitkevich SP, Yurov YB. The phylogeny of human chromosome specific alpha satellites. Chromosoma. 1988;96:443–453. - PubMed
-
- Alkan C, Eichler EE, Bailey JA, Sahinalp SC, Tuzun E. The role of unequal crossover in alpha-satellite DNA evolution: a computational analysis. J Comput Biol. 2004;11:933–944. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

