Orphelia: predicting genes in metagenomic sequencing reads
- PMID: 19429689
- PMCID: PMC2703946
- DOI: 10.1093/nar/gkp327
Orphelia: predicting genes in metagenomic sequencing reads
Abstract
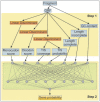
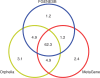
Metagenomic sequencing projects yield numerous sequencing reads of a diverse range of uncultivated and mostly yet unknown microorganisms. In many cases, these sequencing reads cannot be assembled into longer contigs. Thus, gene prediction tools that were originally developed for whole-genome analysis are not suitable for processing metagenomes. Orphelia is a program for predicting genes in short DNA sequences that is available through a web server application (http://orphelia.gobics.de). Orphelia utilizes prediction models that were created with machine learning techniques on the basis of a wide range of annotated genomes. In contrast to other methods for metagenomic gene prediction, Orphelia has fragment length-specific prediction models for the two most popular sequencing techniques in metagenomics, chain termination sequencing and pyrosequencing. These models ensure highly specific gene predictions.
Figures

References
-
- Ronaghi M, Uhlén M, Nyreén P. A sequencing method based on real-time pyrophosphate. Science. 1998;281:363–365. - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. - PubMed
-
- Krause L, Diaz NN, Bartels D, Edwards RA, Pühler A, Rohwer F, Meyer F, Stoye J. Finding novel genes in bacterial communities isolated from the environment. Bioinformatics. 2006;22:e281–e289. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

