Functional mapping of the interaction between TDP-43 and hnRNP A2 in vivo
- PMID: 19429692
- PMCID: PMC2709582
- DOI: 10.1093/nar/gkp342
Functional mapping of the interaction between TDP-43 and hnRNP A2 in vivo
Abstract
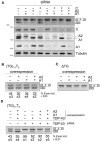
Nuclear factor TDP-43 has been reported to play multiple roles in transcription, pre-mRNA splicing, mRNA stability and mRNA transport. From a structural point of view, TDP-43 is a member of the hnRNP protein family whose structure includes two RRM domains flanked by the N-terminus and C-terminal regions. Like many members of this family, the C-terminal region can interact with cellular factors and thus serve to modulate its function. Previously, we have described that TDP-43 binds to several members of the hnRNP A/B family through this region. In this work, we set up a coupled minigene/siRNA cellular system that allows us to obtain in vivo data to address the functional significance of TDP-43-recruited hnRNP complex formation. Using this method, we have finely mapped the interaction between TDP-43 and the hnRNP A2 protein to the region comprised between amino acid residues 321 and 366. Our results provide novel details of protein-protein interactions in splicing regulation. In addition, we provide further insight on TDP-43 functional properties, particularly the lack of effects, as seen with our assays, of the disease-associated mutations that fall within the TDP-43 321-366 region: Q331K, M337V and G348C.
Figures







References
-
- Buratti E, Baralle FE. Multiple roles of TDP-43 in gene expression, splicing regulation, and human disease. Front. Biosci. 2008;13:867–878. - PubMed
-
- Wang IF, Wu LS, Chang HY, Shen CK. TDP-43, the signature protein of FTLD-U, is a neuronal activity-responsive factor. J. Neurochem. 2008;105:797–806. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

