Role of dimerization and substrate exclusion in the regulation of bone morphogenetic protein-1 and mammalian tolloid
- PMID: 19429706
- PMCID: PMC2689009
- DOI: 10.1073/pnas.0812178106
Role of dimerization and substrate exclusion in the regulation of bone morphogenetic protein-1 and mammalian tolloid
Abstract
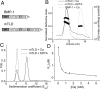
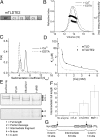
The bone morphogenetic protein (BMP)-1/tolloid metalloproteinases are evolutionarily conserved enzymes that are fundamental to dorsal-ventral patterning and tissue morphogenesis. The lack of knowledge regarding how these proteinases recognize and cleave their substrates represents a major hurdle to understanding tissue assembly and embryonic patterning. Although BMP-1 and mammalian tolloid (mTLD) are splice variants, it is puzzling why BMP-1, which lacks 3 of the 7 noncatalytic domains present in all other family members, is the most effective proteinase. Using a combination of single-particle electron microscopy, small-angle X-ray scattering, and other biophysical measurements in solution, we show that mTLD, but not BMP-1, forms a calcium-ion-dependent dimer under physiological conditions. Using a domain deletion approach, we provide evidence that EGF2, which is absent in BMP-1, is critical to the formation of the dimer. Based on a combination of structural and functional data, we propose that mTLD activity is regulated by a substrate exclusion mechanism. These results provide a mechanistic insight into how alternative splicing of the Bmp1 gene produces 2 proteinases with differing biological activities and have broad implications for regulation of BMP-1/mTLD and related proteinases during BMP signaling and tissue assembly.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Takahara K, Lyons GE, Greenspan DS. Bone morphogenetic protein-1 and a mammalian tolloid homologue (mTld) are encoded by alternatively spliced transcripts which are differentially expressed in some tissues. J Biol Chem. 1994;269:32572–32578. - PubMed
-
- Hojima Y, van der Rest M, Prockop DJ. Type I procollagen carboxyl-terminal proteinase from chick embryo tendons. Purification and characterization. J Biol Chem. 1985;260:15996–16003. - PubMed
-
- Unsold C, Pappano WN, Imamura Y, Steiglitz BM, Greenspan DS. Biosynthetic processing of the pro-alpha 1(V)2pro-alpha 2(V) collagen heterotrimer by bone morphogenetic protein-1 and furin-like proprotein convertases. J Biol Chem. 2002;277:5596–5602. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

