MetaboAnalyst: a web server for metabolomic data analysis and interpretation
- PMID: 19429898
- PMCID: PMC2703878
- DOI: 10.1093/nar/gkp356
MetaboAnalyst: a web server for metabolomic data analysis and interpretation
Abstract
Metabolomics is a newly emerging field of 'omics' research that is concerned with characterizing large numbers of metabolites using NMR, chromatography and mass spectrometry. It is frequently used in biomarker identification and the metabolic profiling of cells, tissues or organisms. The data processing challenges in metabolomics are quite unique and often require specialized (or expensive) data analysis software and a detailed knowledge of cheminformatics, bioinformatics and statistics. In an effort to simplify metabolomic data analysis while at the same time improving user accessibility, we have developed a freely accessible, easy-to-use web server for metabolomic data analysis called MetaboAnalyst. Fundamentally, MetaboAnalyst is a web-based metabolomic data processing tool not unlike many of today's web-based microarray analysis packages. It accepts a variety of input data (NMR peak lists, binned spectra, MS peak lists, compound/concentration data) in a wide variety of formats. It also offers a number of options for metabolomic data processing, data normalization, multivariate statistical analysis, graphing, metabolite identification and pathway mapping. In particular, MetaboAnalyst supports such techniques as: fold change analysis, t-tests, PCA, PLS-DA, hierarchical clustering and a number of more sophisticated statistical or machine learning methods. It also employs a large library of reference spectra to facilitate compound identification from most kinds of input spectra. MetaboAnalyst guides users through a step-by-step analysis pipeline using a variety of menus, information hyperlinks and check boxes. Upon completion, the server generates a detailed report describing each method used, embedded with graphical and tabular outputs. MetaboAnalyst is capable of handling most kinds of metabolomic data and was designed to perform most of the common kinds of metabolomic data analyses. MetaboAnalyst is accessible at http://www.metaboanalyst.ca.
Figures
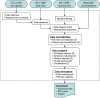

References
-
- Fiehn O. Metabolomics—the link between genotypes and phenotypes. Plant Mol. Biol. 2002;48:155–171. - PubMed
-
- Wishart DS. Quantitative metabolomics using NMR. Trends Anal. Chem. 2008;27:228–237.
-
- Moolenaar SH, Engelke UFH, Wevers RA. Proton nuclear magnetic resonance spectroscopy of body fluids in the field of inborn errors of metabolism. Ann. Clin. Biochem. 2003;40:16–24. - PubMed
-
- Kaddurah-Daouk R, Kristal BS, Weinshilboum RM. Metabolomics: a global biochemical approach to drug response and disease. Ann. Rev. Pharmacol. Toxicol. 2008;48:653–683. - PubMed
-
- Bino RJ, Hall RD, Fiehn O, Kopka J, Saito K, Draper J, Nikolau BJ, Mendes P, Roessner-Tunali U, Beale MH, et al. Potential of metabolomics as a functional genomics tool. Trends Plant Sci. 2004;9:418–425. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

