A short antisense oligonucleotide masking a unique intronic motif prevents skipping of a critical exon in spinal muscular atrophy
- PMID: 19430205
- PMCID: PMC2734876
- DOI: 10.4161/rna.6.3.8723
A short antisense oligonucleotide masking a unique intronic motif prevents skipping of a critical exon in spinal muscular atrophy
Abstract
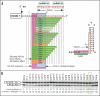
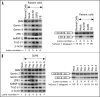
Spinal muscular atrophy (SMA) is the leading genetic cause of infant mortality. Most SMA cases are associated with the low levels of SMN owing to deletion of Survival Motor Neuron 1 (SMN1). SMN2, a nearly identical copy of SMN1, fails to compensate for the loss of SMN1 due to predominant skipping of exon 7. Hence, correction of aberrant splicing of SMN2 exon 7 holds the potential for cure of SMA. Here we report an 8-mer antisense oligonucleotide (ASO) to have a profound stimulatory response on correction of aberrant splicing of SMN2 exon 7 by binding to a unique GC-rich sequence located within intron 7 of SMN2. We confirm that the splicing-switching ability of this short ASO comes with a high degree of specificity and reduced off-target effect compared to larger ASOs targeting the same sequence. We further demonstrate that a single low nanomolar dose of this 8-mer ASO substantially increases the levels of SMN and a host of factors including Gemin 2, Gemin 8, ZPR1, hnRNP Q and Tra2-beta1 known to be down-regulated in SMA. Our findings underscore the advantages and unmatched potential of very short ASOs in splicing modulation in vivo.
Figures



References
-
- Xing Y, Lee C. Relating alternative splicing to proteome complexity and genome evolution. Adv Exp Med Biol. 2007:623:42–9. - PubMed
-
- Nilsen TW. The spliceosome: the most complex macromolecular machine in the cell. Bioessays. 2003;25:1147–9. - PubMed
-
- Matlin AJ, Moore MJ. Spliceosome assembly and composition. Adv Exp Med Biol. 2007;623:14–35. - PubMed
-
- Hertel KJ. Combinatorial control of exon recognition. J Biol Chem. 2008;283:1211–5. - PubMed
-
- Lin S, Fu XD. SR proteins and related factors in alternative splicing. Adv Exp Med Biol. 2007;623:107–22. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
