In silico molecular engineering for a targeted replacement in a tumor-homing peptide
- PMID: 19432404
- PMCID: PMC2734192
- DOI: 10.1021/jp9006119
In silico molecular engineering for a targeted replacement in a tumor-homing peptide
Abstract
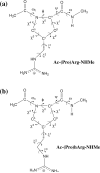
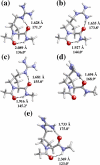
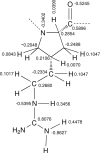
A new amino acid has been designed as a replacement for arginine (Arg, R) to protect the tumor-homing pentapeptide CREKA (Cys-Arg-Glu-Lys-Ala) from proteases. This amino acid, denoted (Pro)hArg, is characterized by a proline skeleton bearing a specifically oriented guanidinium side chain. This residue combines the ability of Pro to induce turn-like conformations with the Arg side-chain functionality. The conformational profile of the CREKA analogue incorporating this Arg substitute has been investigated by a combination of simulated annealing and molecular dynamics. Comparison of the results with those previously obtained for the natural CREKA shows that (Pro)hArg significantly reduces the conformational flexibility of the peptide. Although some changes are observed in the backbone...backbone and side-chain...side-chain interactions, the modified peptide exhibits a strong tendency to accommodate turn conformations centered at the (Pro)hArg residue and the overall shape of the molecule in the lowest energy conformations characterized for the natural and the modified peptides exhibit a high degree of similarity. In particular, the turn orients the backbone such that the Arg, Glu, and Lys side chains face the same side of the molecule, which is considered important for bioactivity. These results suggest that replacement of Arg by (Pro)hArg in CREKA may be useful in providing resistance against proteolytic enzymes while retaining conformational features which are essential for tumor-homing activity.
Figures

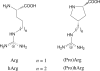

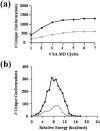


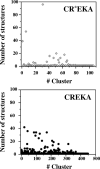
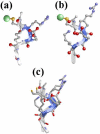
Similar articles
-
Conformational profile of a proline-arginine hybrid.J Chem Inf Model. 2010 Oct 25;50(10):1781-9. doi: 10.1021/ci100135f. J Chem Inf Model. 2010. PMID: 20886854 Free PMC article.
-
Exploring the energy landscape of a molecular engineered analog of a tumor-homing peptide.Phys Chem Chem Phys. 2011 Jun 7;13(21):9986-94. doi: 10.1039/c0cp02572k. Epub 2011 Jan 24. Phys Chem Chem Phys. 2011. PMID: 21258721 Free PMC article.
-
The energy landscape of a selective tumor-homing pentapeptide.J Phys Chem B. 2008 Jul 24;112(29):8692-700. doi: 10.1021/jp711477k. Epub 2008 Jun 28. J Phys Chem B. 2008. PMID: 18588341 Free PMC article.
-
Isomerization of the Xaa-Pro peptide bond induced by ionic interactions of arginine.Biopolymers. 1996 Jun;38(6):673-82. doi: 10.1002/(sici)1097-0282(199606)38:6<673::aid-bip1>3.0.co;2-o. Biopolymers. 1996. PMID: 8652789
-
Influence of the dye presence on the conformational preferences of CREKA, a tumor homing linear pentapeptide.Biopolymers. 2009;92(2):83-93. doi: 10.1002/bip.21122. Biopolymers. 2009. PMID: 19051312 Free PMC article.
Cited by
-
Conformational profile of a proline-arginine hybrid.J Chem Inf Model. 2010 Oct 25;50(10):1781-9. doi: 10.1021/ci100135f. J Chem Inf Model. 2010. PMID: 20886854 Free PMC article.
-
Nanoparticle-induced vascular blockade in human prostate cancer.Blood. 2010 Oct 14;116(15):2847-56. doi: 10.1182/blood-2010-03-274258. Epub 2010 Jun 29. Blood. 2010. PMID: 20587786 Free PMC article.
-
Fibrin-targeted block copolymers for the prevention of postsurgical adhesions.J Biomed Mater Res B Appl Biomater. 2011 Oct;99(1):102-10. doi: 10.1002/jbm.b.31876. Epub 2011 Jun 21. J Biomed Mater Res B Appl Biomater. 2011. PMID: 21695779 Free PMC article.
-
Exploring the energy landscape of a molecular engineered analog of a tumor-homing peptide.Phys Chem Chem Phys. 2011 Jun 7;13(21):9986-94. doi: 10.1039/c0cp02572k. Epub 2011 Jan 24. Phys Chem Chem Phys. 2011. PMID: 21258721 Free PMC article.
-
New p32/gC1qR Ligands for Targeted Tumor Drug Delivery.Chembiochem. 2016 Apr 1;17(7):570-5. doi: 10.1002/cbic.201500564. Epub 2016 Feb 19. Chembiochem. 2016. PMID: 26895508 Free PMC article.
References
-
- Hutchinson JN, Muller WJ. Oncogene. 2000;19:6130. - PubMed
-
- Ruoslahti E. 2007. unpublished results.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

