Biophysical interactions with model lipid membranes: applications in drug discovery and drug delivery
- PMID: 19432455
- PMCID: PMC2757518
- DOI: 10.1021/mp9000662
Biophysical interactions with model lipid membranes: applications in drug discovery and drug delivery
Abstract
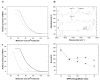
The transport of drugs or drug delivery systems across the cell membrane is a complex biological process, often difficult to understand because of its dynamic nature. In this regard, model lipid membranes, which mimic many aspects of cell-membrane lipids, have been very useful in helping investigators to discern the roles of lipids in cellular interactions. One can use drug-lipid interactions to predict pharmacokinetic properties of drugs, such as their transport, biodistribution, accumulation, and hence efficacy. These interactions can also be used to study the mechanisms of transport, based on the structure and hydrophilicity/hydrophobicity of drug molecules. In recent years, model lipid membranes have also been explored to understand their mechanisms of interactions with peptides, polymers, and nanocarriers. These interaction studies can be used to design and develop efficient drug delivery systems. Changes in the lipid composition of cells and tissue in certain disease conditions may alter biophysical interactions, which could be explored to develop target-specific drugs and drug delivery systems. In this review, we discuss different model membranes, drug-lipid interactions and their significance, studies of model membrane interactions with nanocarriers, and how biophysical interaction studies with lipid model membranes could play an important role in drug discovery and drug delivery.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

