Benchmark experimental data set and assessment of adsorption free energy for peptide-surface interactions
- PMID: 19432493
- PMCID: PMC2756418
- DOI: 10.1021/la8042186
Benchmark experimental data set and assessment of adsorption free energy for peptide-surface interactions
Abstract
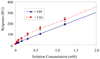
With the increasing interest in protein adsorption in fields ranging from bionanotechnology to biomedical engineering, there is a growing need to understand protein-surface interactions at a fundamental level, such as the interaction between individual amino acid residues of a protein and functional groups presented by a surface. However, relatively little data are available that experimentally provide a quantitative, comparative measure of these types of interactions. To address this deficiency, the objective of this study was to generate a database of experimentally measured standard state adsorption free energy (DeltaGoads) values for a wide variety of amino acid residue-surface interactions using a host-guest peptide and alkanethiol self-assembled monolayers (SAMs) with polymer-like functionality as the model system. The host-guest amino acid sequence was synthesized in the form of TGTG-X-GTGT, where G and T are glycine and threonine amino acid residues and X represents a variable residue. In this paper, we report DeltaGoads values for the adsorption of 12 different types of the host-guest peptides on a set of nine different SAM surfaces, for a total of 108 peptide-surface systems. The DeltaGoads values for these 108 peptide-surface combinations show clear trends in adsorption behavior that are dependent on both peptide composition and surface chemistry. These data provide a benchmark experimental data set from which fundamental interactions that govern peptide and protein adsorption behavior can be better understood and compared.
Figures





References
-
- Hruby VJ, Matsunaga TO. Applications of Synthetic Peptides. In: Grant GA, editor. Synthetic peptides: a user's guide. 2nd ed. Oxford; New York: Oxford University Press; 2002. pp. 292–376.
-
- Zhang W, Laursen RA. Artificial antifreeze polypeptides: alpha-helical peptides with KAAK motifs have antifreeze and ice crystal morphology modifying properties. FEBS Lett. 1999;455(3):372–376. - PubMed
-
- Corradin G, Spertini F, Verdini A. Medicinal application of long synthetic peptide technology. Expert Opinion on Biological Therapy. 2004;4(10):1629–1639. - PubMed
-
- Srivastava AK, Castillo G, Wergedal JE, Mohan S, Baylink DJ. Development and Application of a Synthetic Peptide-Based Osteocalcin Assay for the Measurement of Bone Formation in Mouse Serum. Calcified Tissue International. 2000;67(3):255. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

