Characterization of Vitis vinifera NPR1 homologs involved in the regulation of pathogenesis-related gene expression
- PMID: 19432948
- PMCID: PMC2686700
- DOI: 10.1186/1471-2229-9-54
Characterization of Vitis vinifera NPR1 homologs involved in the regulation of pathogenesis-related gene expression
Abstract
Background: Grapevine protection against diseases needs alternative strategies to the use of phytochemicals, implying a thorough knowledge of innate defense mechanisms. However, signalling pathways and regulatory elements leading to induction of defense responses have yet to be characterized in this species. In order to study defense response signalling to pathogens in Vitis vinifera, we took advantage of its recently completed genome sequence to characterize two putative orthologs of NPR1, a key player in salicylic acid (SA)-mediated resistance to biotrophic pathogens in Arabidopsis thaliana.
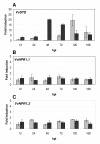
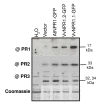
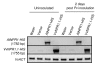
Results: Two cDNAs named VvNPR1.1 and VvNPR1.2 were isolated from Vitis vinifera cv chardonnay, encoding proteins showing 55% and 40% identity to Arabidopsis NPR1 respectively. Constitutive expression of VvNPR1.1 and VvNPR1.2 monitored in leaves of V. vinifera cv chardonnay was found to be enhanced by treatment with benzothiadiazole, a SA analog. In contrast, VvNPR1.1 and VvNPR1.2 transcript levels were not affected during infection of resistant Vitis riparia or susceptible V. vinifera with Plasmopara viticola, the causal agent of downy mildew, suggesting regulation of VvNPR1 activity at the protein level. VvNPR1.1-GFP and VvNPR1.2-GFP fusion proteins were transiently expressed by agroinfiltration in Nicotiana benthamiana leaves, where they localized predominantly to the nucleus. In this system, VvNPR1.1 and VvNPR1.2 expression was sufficient to trigger the accumulation of acidic SA-dependent pathogenesis-related proteins PR1 and PR2, but not of basic chitinases (PR3) in the absence of pathogen infection. Interestingly, when VvNPR1.1 or AtNPR1 were transiently overexpressed in Vitis vinifera leaves, the induction of grapevine PR1 was significantly enhanced in response to P. viticola.
Conclusion: In conclusion, our data identified grapevine homologs of NPR1, and their functional analysis showed that VvNPR1.1 and VvNPR1.2 likely control the expression of SA-dependent defense genes. Overexpression of VvNPR1 has thus the potential to enhance grapevine defensive capabilities upon fungal infection. As a consequence, manipulating VvNPR1 and other signalling elements could open ways to strengthen disease resistance mechanisms in this crop species.
Figures




Similar articles
-
Vitis vinifera VvNPR1.1 is the functional ortholog of AtNPR1 and its overexpression in grapevine triggers constitutive activation of PR genes and enhanced resistance to powdery mildew.Planta. 2011 Aug;234(2):405-17. doi: 10.1007/s00425-011-1412-1. Epub 2011 Apr 20. Planta. 2011. PMID: 21505863
-
General and species-specific transcriptional responses to downy mildew infection in a susceptible (Vitis vinifera) and a resistant (V. riparia) grapevine species.BMC Genomics. 2010 Feb 18;11:117. doi: 10.1186/1471-2164-11-117. BMC Genomics. 2010. PMID: 20167053 Free PMC article.
-
Low level of polymorphism in two putative NPR1 homologs in the Vitaceae family.Biol Direct. 2010 Feb 5;5:9. doi: 10.1186/1745-6150-5-9. Biol Direct. 2010. PMID: 20137081 Free PMC article.
-
The pathogenicity of Plasmopara viticola: a review of evolutionary dynamics, infection strategies and effector molecules.BMC Plant Biol. 2024 Apr 24;24(1):327. doi: 10.1186/s12870-024-05037-0. BMC Plant Biol. 2024. PMID: 38658826 Free PMC article. Review.
-
Advances in understanding grapevine downy mildew: From pathogen infection to disease management.Mol Plant Pathol. 2024 Jan;25(1):e13401. doi: 10.1111/mpp.13401. Epub 2023 Nov 22. Mol Plant Pathol. 2024. PMID: 37991155 Free PMC article. Review.
Cited by
-
Transcriptomic analysis of Chinese wild Vitis pseudoreticulata in response to Plasmopara viticola.Protoplasma. 2019 Sep;256(5):1409-1424. doi: 10.1007/s00709-019-01387-x. Epub 2019 May 21. Protoplasma. 2019. PMID: 31115695
-
CRISPR/Cas9-driven double modification of grapevine MLO6-7 imparts powdery mildew resistance, while editing of NPR3 augments powdery and downy mildew tolerance.Plant J. 2025 Apr;122(2):e17204. doi: 10.1111/tpj.17204. Epub 2024 Dec 8. Plant J. 2025. PMID: 39645650 Free PMC article.
-
Biologically active and antimicrobial peptides from plants.Biomed Res Int. 2015;2015:102129. doi: 10.1155/2015/102129. Epub 2015 Mar 1. Biomed Res Int. 2015. PMID: 25815307 Free PMC article. Review.
-
Berry flesh and skin ripening features in Vitis vinifera as assessed by transcriptional profiling.PLoS One. 2012;7(6):e39547. doi: 10.1371/journal.pone.0039547. Epub 2012 Jun 29. PLoS One. 2012. PMID: 22768087 Free PMC article.
-
Identification of a strawberry NPR-like gene involved in negative regulation of the salicylic acid-mediated defense pathway.PLoS One. 2018 Oct 12;13(10):e0205790. doi: 10.1371/journal.pone.0205790. eCollection 2018. PLoS One. 2018. PMID: 30312354 Free PMC article.
References
-
- Thomma BP, Eggermont K, Penninckx IA, Mauch-Mani B, Vogelsang R, Cammue B, Broekaert WF. Separate jasmonate-dependent and salicylate- dependent defense response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci USA. 1998;95:15107–15111. doi: 10.1073/pnas.95.25.15107. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

