Insights into the regulation of intrinsically disordered proteins in the human proteome by analyzing sequence and gene expression data
- PMID: 19432952
- PMCID: PMC2718516
- DOI: 10.1186/gb-2009-10-5-r50
Insights into the regulation of intrinsically disordered proteins in the human proteome by analyzing sequence and gene expression data
Abstract
Background: Disordered proteins need to be expressed to carry out specified functions; however, their accumulation in the cell can potentially cause major problems through protein misfolding and aggregation. Gene expression levels, mRNA decay rates, microRNA (miRNA) targeting and ubiquitination have critical roles in the degradation and disposal of human proteins and transcripts. Here, we describe a study examining these features to gain insights into the regulation of disordered proteins.
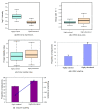
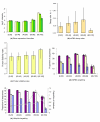
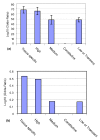
Results: In comparison with ordered proteins, disordered proteins have a greater proportion of predicted ubiquitination sites. The transcripts encoding disordered proteins also have higher proportions of predicted miRNA target sites and higher mRNA decay rates, both of which are indicative of the observed lower gene expression levels. The results suggest that the disordered proteins and their transcripts are present in the cell at low levels and/or for a short time before being targeted for disposal. Surprisingly, we find that for a significant proportion of highly disordered proteins, all four of these trends are reversed. Predicted estimates for miRNA targets, ubiquitination and mRNA decay rate are low in the highly disordered proteins that are constitutively and/or highly expressed.
Conclusions: Mechanisms are in place to protect the cell from these potentially dangerous proteins. The evidence suggests that the enrichment of signals for miRNA targeting and ubiquitination may help prevent the accumulation of disordered proteins in the cell. Our data also provide evidence for a mechanism by which a significant proportion of highly disordered proteins (with high expression levels) can escape rapid degradation to allow them to successfully carry out their function.
Figures

References
-
- Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang C, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner EC, Obradovic Z. Intrinsically disordered protein. J Mol Graph Model. 2001;19:26–59. doi: 10.1016/S1093-3263(00)00138-8. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

