MicroRNA signatures predict oestrogen receptor, progesterone receptor and HER2/neu receptor status in breast cancer
- PMID: 19432961
- PMCID: PMC2716495
- DOI: 10.1186/bcr2257
MicroRNA signatures predict oestrogen receptor, progesterone receptor and HER2/neu receptor status in breast cancer
Abstract
Introduction: Breast cancer is a heterogeneous disease encompassing a number of phenotypically diverse tumours. Expression levels of the oestrogen, progesterone and HER2/neu receptors which characterize clinically distinct breast tumours have been shown to change during disease progression and in response to systemic therapies. Mi(cro)RNAs play critical roles in diverse biological processes and are aberrantly expressed in several human neoplasms including breast cancer, where they function as regulators of tumour behaviour and progression. The aims of this study were to identify miRNA signatures that accurately predict the oestrogen receptor (ER), progesterone receptor (PR) and HER2/neu receptor status of breast cancer patients to provide insight into the regulation of breast cancer phenotypes and progression.
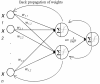
Methods: Expression profiling of 453 miRNAs was performed in 29 early-stage breast cancer specimens. miRNA signatures associated with ER, PR and HER2/neu status were generated using artificial neural networks (ANN), and expression of specific miRNAs was validated using RQ-PCR.
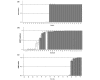
Results: Stepwise ANN analysis identified predictive miRNA signatures corresponding with oestrogen (miR-342, miR-299, miR-217, miR-190, miR-135b, miR-218), progesterone (miR-520g, miR-377, miR-527-518a, miR-520f-520c) and HER2/neu (miR-520d, miR-181c, miR-302c, miR-376b, miR-30e) receptor status. MiR-342 and miR-520g expression was further analysed in 95 breast tumours. MiR-342 expression was highest in ER and HER2/neu-positive luminal B tumours and lowest in triple-negative tumours. MiR-520g expression was elevated in ER and PR-negative tumours.
Conclusions: This study demonstrates that ANN analysis reliably identifies biologically relevant miRNAs associated with specific breast cancer phenotypes. The association of specific miRNAs with ER, PR and HER2/neu status indicates a role for these miRNAs in disease classification of breast cancer. Decreased expression of miR-342 in the therapeutically challenging triple-negative breast tumours, increased miR-342 expression in the luminal B tumours, and downregulated miR-520g in ER and PR-positive tumours indicates that not only is dysregulated miRNA expression a marker for poorer prognosis breast cancer, but that it could also present an attractive target for therapeutic intervention.
Figures





Similar articles
-
Identification and validation of oncologic miRNA biomarkers for luminal A-like breast cancer.PLoS One. 2014 Jan 31;9(1):e87032. doi: 10.1371/journal.pone.0087032. eCollection 2014. PLoS One. 2014. PMID: 24498016 Free PMC article.
-
Coordinated expression of ER, PR and HER2 define different prognostic subtypes among poorly differentiated breast carcinomas.Histopathology. 2009 Sep;55(3):346-52. doi: 10.1111/j.1365-2559.2009.03380.x. Histopathology. 2009. PMID: 19723150
-
Correlation of miRNA expression profiling in surgical pathology materials, with Ki-67, HER2, ER and PR in breast cancer patients.Int J Biol Markers. 2015 May 26;30(2):e190-9. doi: 10.5301/jbm.5000141. Int J Biol Markers. 2015. PMID: 25907662
-
Estrogen, progesterone, and HER2/neu receptor discordance between primary and metastatic breast tumours-a review.Cancer Metastasis Rev. 2016 Sep;35(3):427-37. doi: 10.1007/s10555-016-9631-3. Cancer Metastasis Rev. 2016. PMID: 27405651 Review.
-
From microRNA functions to microRNA therapeutics: novel targets and novel drugs in breast cancer research and treatment (Review).Int J Oncol. 2013 Oct;43(4):985-94. doi: 10.3892/ijo.2013.2059. Epub 2013 Aug 12. Int J Oncol. 2013. PMID: 23939688 Free PMC article. Review.
Cited by
-
Prospective Assessment of Systemic MicroRNAs as Markers of Response to Neoadjuvant Chemotherapy in Breast Cancer.Cancers (Basel). 2020 Jul 7;12(7):1820. doi: 10.3390/cancers12071820. Cancers (Basel). 2020. PMID: 32645898 Free PMC article.
-
Prognostic Implications of MicroRNA-21 Overexpression in Invasive Ductal Carcinomas of the Breast.J Breast Cancer. 2011 Dec;14(4):269-75. doi: 10.4048/jbc.2011.14.4.269. Epub 2011 Dec 27. J Breast Cancer. 2011. PMID: 22323912 Free PMC article.
-
Modulatory role of miRNAs in thyroid and breast cancer progression and insights into their therapeutic manipulation.Curr Res Pharmacol Drug Discov. 2022 Oct 3;3:100131. doi: 10.1016/j.crphar.2022.100131. eCollection 2022. Curr Res Pharmacol Drug Discov. 2022. PMID: 36568259 Free PMC article. Review.
-
Analysis of microRNAs in Exosomes of Breast Cancer Patients in Search of Molecular Prognostic Factors in Brain Metastases.Int J Mol Sci. 2022 Mar 27;23(7):3683. doi: 10.3390/ijms23073683. Int J Mol Sci. 2022. PMID: 35409043 Free PMC article.
-
MicroRNAs Contribute to Breast Cancer Invasiveness.Cells. 2019 Oct 31;8(11):1361. doi: 10.3390/cells8111361. Cells. 2019. PMID: 31683635 Free PMC article. Review.
References
-
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, Rijn M van de, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lonning P, Borresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. - DOI - PMC - PubMed
-
- Potti A, Dressman HK, Bild A, Riedel RF, Chan G, Sayer R, Cragun J, Cottrill H, Kelley MJ, Petersen R, Harpole D, Marks J, Berchuck A, Ginsburg GS, Febbo P, Lancaster J, Nevins JR. Genomic signatures to guide the use of chemotherapeutics. Nat Med. 2006;12:1294–1300. doi: 10.1038/nm1491. - DOI - PubMed
-
- Chin SF, Teschendorff AE, Marioni JC, Wang Y, Barbosa-Morais NL, Thorne NP, Costa JL, Pinder SE, Wiel MA van de, Green AR, Ellis IO, Porter PL, Tavaré S, Brenton JD, Ylstra B, Caldas C. High-resolution aCGH and expression profiling identifies a novel genomic subtype of ER negative breast cancer. Genome Biol. 2007;8:R215. doi: 10.1186/gb-2007-8-10-r215. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

