Estimating parameters for generalized mass action models with connectivity information
- PMID: 19432964
- PMCID: PMC2694188
- DOI: 10.1186/1471-2105-10-140
Estimating parameters for generalized mass action models with connectivity information
Abstract
Background: Determining the parameters of a mathematical model from quantitative measurements is the main bottleneck of modelling biological systems. Parameter values can be estimated from steady-state data or from dynamic data. The nature of suitable data for these two types of estimation is rather different. For instance, estimations of parameter values in pathway models, such as kinetic orders, rate constants, flux control coefficients or elasticities, from steady-state data are generally based on experiments that measure how a biochemical system responds to small perturbations around the steady state. In contrast, parameter estimation from dynamic data requires time series measurements for all dependent variables. Almost no literature has so far discussed the combined use of both steady-state and transient data for estimating parameter values of biochemical systems.
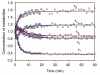
Results: In this study we introduce a constrained optimization method for estimating parameter values of biochemical pathway models using steady-state information and transient measurements. The constraints are derived from the flux connectivity relationships of the system at the steady state. Two case studies demonstrate the estimation results with and without flux connectivity constraints. The unconstrained optimal estimates from dynamic data may fit the experiments well, but they do not necessarily maintain the connectivity relationships. As a consequence, individual fluxes may be misrepresented, which may cause problems in later extrapolations. By contrast, the constrained estimation accounting for flux connectivity information reduces this misrepresentation and thereby yields improved model parameters.
Conclusion: The method combines transient metabolic profiles and steady-state information and leads to the formulation of an inverse parameter estimation task as a constrained optimization problem. Parameter estimation and model selection are simultaneously carried out on the constrained optimization problem and yield realistic model parameters that are more likely to hold up in extrapolations with the model.
Figures

Similar articles
-
Extraction of elementary rate constants from global network analysis of E. coli central metabolism.BMC Syst Biol. 2008 May 7;2:41. doi: 10.1186/1752-0509-2-41. BMC Syst Biol. 2008. PMID: 18462493 Free PMC article.
-
An improved hybrid of particle swarm optimization and the gravitational search algorithm to produce a kinetic parameter estimation of aspartate biochemical pathways.Biosystems. 2017 Dec;162:81-89. doi: 10.1016/j.biosystems.2017.09.013. Epub 2017 Sep 23. Biosystems. 2017. PMID: 28951204
-
A method for estimation of elasticities in metabolic networks using steady state and dynamic metabolomics data and linlog kinetics.BMC Bioinformatics. 2006 Dec 21;7:540. doi: 10.1186/1471-2105-7-540. BMC Bioinformatics. 2006. PMID: 17184531 Free PMC article.
-
Recent developments in parameter estimation and structure identification of biochemical and genomic systems.Math Biosci. 2009 Jun;219(2):57-83. doi: 10.1016/j.mbs.2009.03.002. Epub 2009 Mar 25. Math Biosci. 2009. PMID: 19327372 Free PMC article. Review.
-
Applications of model simulation in pharmacological fields and the problems of theoretical reliability.Drug Metab Pharmacokinet. 2024 Jun;56:100996. doi: 10.1016/j.dmpk.2024.100996. Epub 2024 Jan 4. Drug Metab Pharmacokinet. 2024. PMID: 38797090 Review.
Cited by
-
VEGFR1 promotes cell migration and proliferation through PLCγ and PI3K pathways.NPJ Syst Biol Appl. 2017 Dec 19;4:1. doi: 10.1038/s41540-017-0037-9. eCollection 2018. NPJ Syst Biol Appl. 2017. PMID: 29263797 Free PMC article.
-
The best models of metabolism.Wiley Interdiscip Rev Syst Biol Med. 2017 Nov;9(6):10.1002/wsbm.1391. doi: 10.1002/wsbm.1391. Epub 2017 May 19. Wiley Interdiscip Rev Syst Biol Med. 2017. PMID: 28544810 Free PMC article. Review.
-
GA-based Design Algorithms for the Robust Synthetic Genetic Oscillators with Prescribed Amplitude, Period and Phase.Gene Regul Syst Bio. 2010 May 24;4:35-52. doi: 10.4137/grsb.s4818. Gene Regul Syst Bio. 2010. PMID: 20535234 Free PMC article.
-
Comprehensive Review of Models and Methods for Inferences in Bio-Chemical Reaction Networks.Front Genet. 2019 Jun 14;10:549. doi: 10.3389/fgene.2019.00549. eCollection 2019. Front Genet. 2019. PMID: 31258548 Free PMC article. Review.
References
-
- Polisetty PK, Voit EO, Gatzke EP. Proceedings of Foundations of Systems Biology in Engineering. Santa Barbara, CA; 2005. Yield optimization of Saccharomyces cerevisiae using a GMA model and a MILP-based piecewise linear relaxation method.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

