Evaluation of high efficiency gene knockout strategies for Trypanosoma cruzi
- PMID: 19432966
- PMCID: PMC2688506
- DOI: 10.1186/1471-2180-9-90
Evaluation of high efficiency gene knockout strategies for Trypanosoma cruzi
Abstract
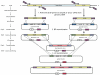
Background: Trypanosoma cruzi, a kinetoplastid protozoan parasite that causes Chagas disease, infects approximately 15 million people in Central and South America. In contrast to the substantial in silico studies of the T. cruzi genome, transcriptome, and proteome, only a few genes have been experimentally characterized and validated, mainly due to the lack of facile methods for gene manipulation needed for reverse genetic studies. Current strategies for gene disruption in T. cruzi are tedious and time consuming. In this study we have compared the conventional multi-step cloning technique with two knockout strategies that have been proven to work in other organisms, one-step-PCR- and Multisite Gateway-based systems.
Results: While the one-step-PCR strategy was found to be the fastest method for production of knockout constructs, it does not efficiently target genes of interest using gene-specific sequences of less than 80 nucleotides. Alternatively, the Multisite Gateway based approach is less time-consuming than conventional methods and is able to efficiently and reproducibly delete target genes.
Conclusion: Using the Multisite Gateway strategy, we have rapidly produced constructs that successfully produce specific gene deletions in epimastigotes of T. cruzi. This methodology should greatly facilitate reverse genetic studies in T. cruzi.
Figures

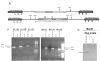


References
-
- Barrett MP, Burchmore RJ, Stich A, Lazzari JO, Frasch AC, Cazzulo JJ, Krishna S. The trypanosomiases. Lancet. 2003;362(9394):1469–1480. - PubMed
-
- Control of Chagas disease. World Health Organ Tech Rep Ser. 2002;905:i–iv. 1–109, back cover. - PubMed
-
- TDR/PAHO/WHO Scientific Working Group Report. Reporte sobre la enfermedad de Chagas. 2007. http://www.who.int/tdr/svc/publications/tdr-research-publications/report...
-
- Tyler KM, Engman DM. The life cycle of Trypanosoma cruzi revisited. Int J Parasitol. 2001;31(5–6):472–481. - PubMed
-
- El-Sayed NM, Myler PJ, Bartholomeu DC, Nilsson D, Aggarwal G, Tran A-N, Ghedin E, Worthey EA, Delcher AL, Blandin G. The Genome Sequence of Trypanosoma cruzi, Etiologic Agent of Chagas Disease. Science. 2005;309(5733):409–415. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

