Assessment of methods and analysis of outcomes for comprehensive optimization of nucleofection
- PMID: 19432988
- PMCID: PMC2683797
- DOI: 10.1186/1479-0556-7-6
Assessment of methods and analysis of outcomes for comprehensive optimization of nucleofection
Abstract
Background: Nucleofection is an emerging technology for delivery of nucleic acids into both the cytoplasm and nucleus of eukaryotic cells with high efficiency. This makes it an ideal technology for gene delivery and siRNA applications. A 96-well format has recently been made available for high-throughput nucleofection, however conditions must be optimized for delivery into each specific cell type. Screening each 96-well plate can be expensive, and descriptions of methods and outcomes to determine the best conditions are lacking in the literature. Here we employ simple methods, including cell counting, microscopy, viability and cytotoxicity assays to describe the minimal experimental methods required to optimize nucleofection conditions for a given cell line.
Methods: We comprehensively measured and analyzed the outcomes of the 96-well nucleofection of pmaxGFP plasmids encoding green fluorescent protein (GFP) into the A-549 human lung epithelial cell line. Fluorescent microscopy and a plate reader were used to respectively observe and quantify green fluorescence in both whole and lysed cells. Cell viability was determined by direct counting/permeability assays, and by both absorbance and fluorescence-based plate reader cytotoxicity assays. Finally, an optimal nucleofection condition was used to deliver siRNA and gene specific knock-down was demonstrated.
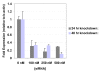
Results: GFP fluorescence among conditions ranged from non-existent to bright, based upon the fluorescent microscopy and plate reader results. Correlation between direct counting of cells and plate-based cytotoxicity assays were from R = .81 to R = .88, depending on the assay. Correlation between the GFP fluorescence of lysed and unlysed cells was high, ranging from R = .91 to R = .97. Finally, delivery of a pooled sample of siRNAs targeting the gene relA using an optimized nucleofection condition resulted in a 70-95% knock down of the gene over 48 h with 90-97% cell viability.
Conclusion: Our results show the optimal 96-well nucleofection conditions for the widely-used human cell line, A-549. We describe simple, effective methods for determining optimal conditions with high confidence, providing a useful road map for other laboratories planning optimization of specific cell lines or primary cells. Our analysis of outcomes suggests the need to only measure unlysed, whole-cell fluorescence and cell metabolic activity using a plate reader cytotoxicity assay to determine the best conditions for 96-well nucleofection.
Figures


Similar articles
-
The two hit hypothesis: an improved method for siRNA-mediated gene silencing in stimulated primary human T cells.J Immunol Methods. 2013 Oct 31;396(1-2):116-27. doi: 10.1016/j.jim.2013.08.005. Epub 2013 Aug 27. J Immunol Methods. 2013. PMID: 23988722
-
Nucleofection is a highly effective gene transfer technique for human melanoma cell lines.Exp Dermatol. 2008 May;17(5):405-11. doi: 10.1111/j.1600-0625.2007.00687.x. Epub 2008 Feb 27. Exp Dermatol. 2008. PMID: 18312380
-
High transfection efficiency of porcine peripheral blood T cells via nucleofection.Vet Immunol Immunopathol. 2011 Dec 15;144(3-4):179-86. doi: 10.1016/j.vetimm.2011.10.003. Epub 2011 Oct 15. Vet Immunol Immunopathol. 2011. PMID: 22055481
-
Efficient non-viral transfection of primary human adult chondrocytes in a high-throughput format.Osteoarthritis Cartilage. 2009 Jun;17(6):813-7. doi: 10.1016/j.joca.2008.11.004. Epub 2008 Nov 13. Osteoarthritis Cartilage. 2009. PMID: 19056302
-
Nucleic acid direct delivery to fibroblasts: a review of nucleofection and applications.J Biol Eng. 2022 Nov 4;16(1):30. doi: 10.1186/s13036-022-00309-5. J Biol Eng. 2022. PMID: 36329479 Free PMC article. Review.
Cited by
-
Physical non-viral gene delivery methods for tissue engineering.Ann Biomed Eng. 2013 Mar;41(3):446-68. doi: 10.1007/s10439-012-0678-1. Epub 2012 Oct 26. Ann Biomed Eng. 2013. PMID: 23099792 Free PMC article. Review.
-
piggyBac transposon plus insulators overcome epigenetic silencing to provide for stable signaling pathway reporter cell lines.PLoS One. 2013 Dec 20;8(12):e85494. doi: 10.1371/journal.pone.0085494. eCollection 2013. PLoS One. 2013. PMID: 24376882 Free PMC article.
-
SREBP-1 Mediates Angiotensin II-Induced TGF-β1 Upregulation and Glomerular Fibrosis.J Am Soc Nephrol. 2015 Aug;26(8):1839-54. doi: 10.1681/ASN.2013121332. Epub 2014 Nov 14. J Am Soc Nephrol. 2015. PMID: 25398788 Free PMC article.
-
Cytotoxic Activity and Memory T Cell Subset Distribution of in vitro-Stimulated CD8+ T Cells Specific for HER2/neu Epitopes.Front Immunol. 2019 May 9;10:1017. doi: 10.3389/fimmu.2019.01017. eCollection 2019. Front Immunol. 2019. PMID: 31143180 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

